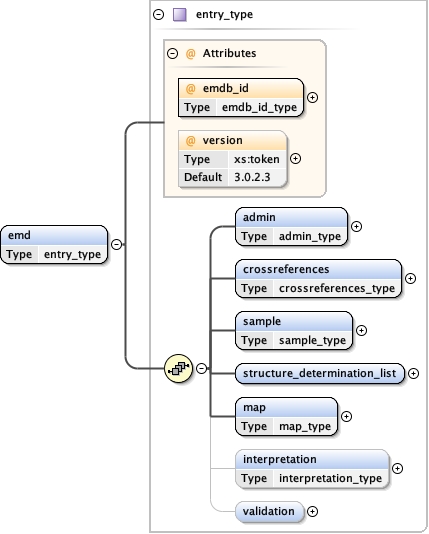
Main schema emdb.xsd
Element emd
Namespace No namespace
Diagram
Type entry_type
Properties
Model
Children admin crossreferences interpretation map sample structure_determination_list validation
Instance
<emd emdb_id= "" version= "3.0.2.3" >
<admin > {1,1} </admin>
<crossreferences > {1,1} </crossreferences>
<sample > {1,1} </sample>
<structure_determination_list > {1,1} </structure_determination_list>
<map format= "CCP4" size_kbytes= "" > {1,1} </map>
<interpretation > {0,1} </interpretation>
<validation > {0,1} </validation>
</emd>
Attributes
Source
<xs:element name= "emd" type= "entry_type" />
Namespace No namespace
Diagram
Type admin_type
Properties
Model
status_history_list{0,1} ,
current_status ,
sites ,
key_dates ,
obsolete_list{0,1} ,
superseded_by_list{0,1} ,
grant_support{0,1} ,
microscopy_center{0,1} ,
contact_author* ,
title ,
authors_list ,
details{0,1} ,
keywords{0,1} ,
replace_existing_entry{0,1}
Children authors_list contact_author current_status details grant_support key_dates keywords microscopy_center obsolete_list replace_existing_entry sites status_history_list superseded_by_list title
Instance
<admin >
<status_history_list > {0,1} </status_history_list>
<current_status > {1,1} </current_status>
<sites > {1,1} </sites>
<key_dates > {1,1} </key_dates>
<obsolete_list > {0,1} </obsolete_list>
<superseded_by_list > {0,1} </superseded_by_list>
<grant_support > {0,1} </grant_support>
<microscopy_center > {0,1} </microscopy_center>
<contact_author private= "true" > {0,unbounded} </contact_author>
<title > {1,1} </title>
<authors_list > {1,1} </authors_list>
<details > {0,1} </details>
<keywords > {0,1} </keywords>
<replace_existing_entry > {0,1} </replace_existing_entry>
</admin>
Source
<xs:element name= "admin" type= "admin_type" />
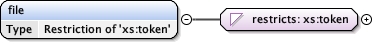
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
PDBe
enumeration
RCSB
enumeration
PDBj
enumeration
PDBc
Source
<xs:element name= "processing_site" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PDBe" />
<xs:enumeration value= "RCSB" />
<xs:enumeration value= "PDBj" />
<xs:enumeration value= "PDBc" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
PDBe
enumeration
PDBj
enumeration
RCSB
enumeration
PDBc
Source
<xs:element name= "deposition" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PDBe" />
<xs:enumeration value= "PDBj" />
<xs:enumeration value= "RCSB" />
<xs:enumeration value= "PDBc" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
PDBe
enumeration
PDBj
enumeration
RCSB
enumeration
PDBc
Source
<xs:element name= "last_processing" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PDBe" />
<xs:enumeration value= "PDBj" />
<xs:enumeration value= "RCSB" />
<xs:enumeration value= "PDBc" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type xs:date
Properties
Source
Namespace No namespace
Diagram
Type extension of contact_details_type
Type hierarchy
Properties
Model
Children country email fax first_name last_name middle_name organization post_or_zip_code role state_or_province street telephone title town_or_city
Instance
Attributes
Source
Element citation_type
Element supramolecule
Namespace No namespace
Diagram
Type base_supramolecule_type
Properties
content
complex
abstract
true
Substitution Group
Used by
Model
name ,
category{0,1} ,
parent ,
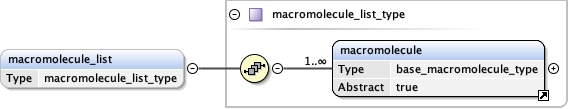
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
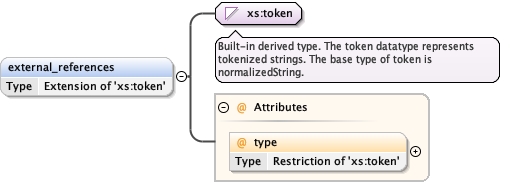
external_references* ,
recombinant_exp_flag{0,1}
Children category details external_references macromolecule_list name number_of_copies oligomeric_state parent recombinant_exp_flag
Instance
<supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
</supramolecule>
Attributes
Source
<xs:element name= "supramolecule" type= "base_supramolecule_type" abstract= "true" />
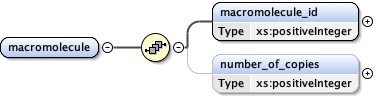
Element macromolecule
Namespace No namespace
Diagram
Type base_macromolecule_type
Properties
content
complex
abstract
true
Substitution Group
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1}
Children details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag
Instance
<macromolecule chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
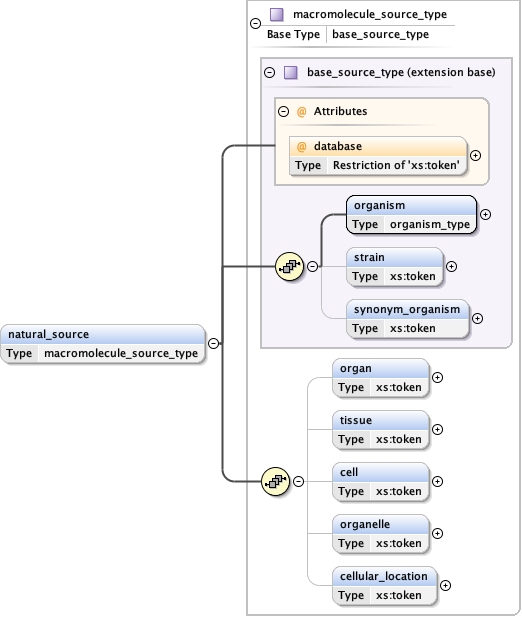
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
</macromolecule>
Attributes
Source
<xs:element name= "macromolecule" type= "base_macromolecule_type" abstract= "true" />
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<natural_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</natural_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "natural_source" type= "macromolecule_source_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type structure_determination_type
Properties
content
complex
maxOccurs
unbounded
Model
Children aggregation_state image_processing macromolecules_and_complexes method microscopy_list specimen_preparation_list
Instance
<structure_determination structure_determination_id= "" >
<method > {1,1} </method>
<aggregation_state > {1,1} </aggregation_state>
<macromolecules_and_complexes > {0,1} </macromolecules_and_complexes>
<specimen_preparation_list > {1,1} </specimen_preparation_list>
<microscopy_list > {1,1} </microscopy_list>
<image_processing image_processing_id= "" > {1,unbounded} </image_processing>
</structure_determination>
Attributes
Source
<xs:element name= "structure_determination" type= "structure_determination_type" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
singleParticle
enumeration
subtomogramAveraging
enumeration
tomography
enumeration
electronCrystallography
enumeration
helical
Source
<xs:element name= "method" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "singleParticle" />
<xs:enumeration value= "subtomogramAveraging" />
<xs:enumeration value= "tomography" />
<xs:enumeration value= "electronCrystallography" />
<xs:enumeration value= "helical" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
particle
enumeration
filament
enumeration
twoDArray
enumeration
threeDArray
enumeration
helicalArray
enumeration
cell
enumeration
tissue
Source
<xs:element name= "aggregation_state" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "particle" />
<xs:enumeration value= "filament" />
<xs:enumeration value= "twoDArray" />
<xs:enumeration value= "threeDArray" />
<xs:enumeration value= "helicalArray" />
<xs:enumeration value= "cell" />
<xs:enumeration value= "tissue" />
</xs:restriction>
</xs:simpleType>
</xs:element>
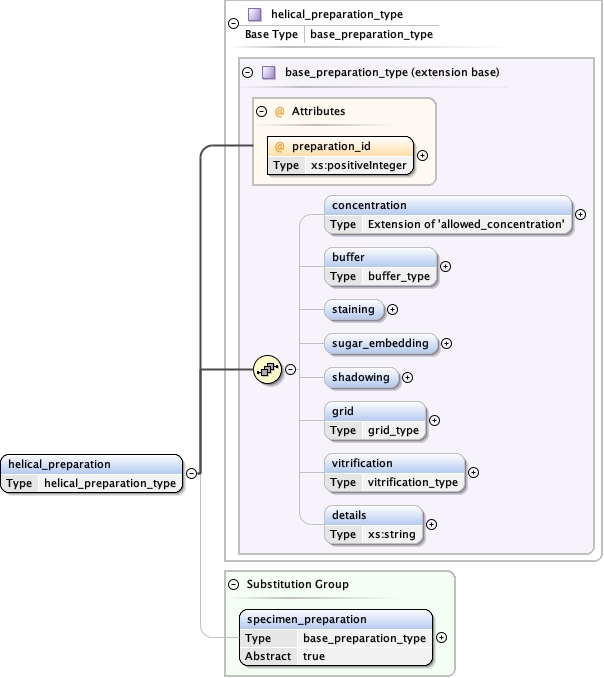
Element specimen_preparation
Namespace No namespace
Diagram
Type base_preparation_type
Properties
content
complex
abstract
true
Substitution Group
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Instance
<specimen_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
</specimen_preparation>
Attributes
Source
<xs:element name= "specimen_preparation" type= "base_preparation_type" abstract= "true" />
Namespace No namespace
Diagram
Type grid_type
Properties
content
complex
minOccurs
0
Model
model{0,1} ,
material{0,1} ,
mesh{0,1} ,
support_film* ,
pretreatment{0,1} ,
details{0,1}
Children details material mesh model pretreatment support_film
Instance
<grid >
<model > {0,1} </model>
<material > {0,1} </material>
<mesh > {0,1} </mesh>
<support_film film_type_id= "" > {0,unbounded} </support_film>
<pretreatment > {0,1} </pretreatment>
<details > {0,1} </details>
</grid>
Source
<xs:element name= "grid" type= "grid_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
COPPER
enumeration
COPPER/PALLADIUM
enumeration
COPPER/RHODIUM
enumeration
GOLD
enumeration
GRAPHENE OXIDE
enumeration
MOLYBDENUM
enumeration
NICKEL
enumeration
NICKEL/TITANIUM
enumeration
PLATINUM
enumeration
SILICON NITRIDE
enumeration
TITANIUM
enumeration
TUNGSTEN
Source
<xs:element name= "material" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "COPPER" />
<xs:enumeration value= "COPPER/PALLADIUM" />
<xs:enumeration value= "COPPER/RHODIUM" />
<xs:enumeration value= "GOLD" />
<xs:enumeration value= "GRAPHENE OXIDE" />
<xs:enumeration value= "MOLYBDENUM" />
<xs:enumeration value= "NICKEL" />
<xs:enumeration value= "NICKEL/TITANIUM" />
<xs:enumeration value= "PLATINUM" />
<xs:enumeration value= "SILICON NITRIDE" />
<xs:enumeration value= "TITANIUM" />
<xs:enumeration value= "TUNGSTEN" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
CARBON
enumeration
CELLULOSE ACETATE
enumeration
FORMVAR
enumeration
GOLD
enumeration
GRAPHENE
enumeration
GRAPHENE OXIDE
enumeration
PARLODION
Source
<xs:element name= "film_material" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "CARBON" />
<xs:enumeration value= "CELLULOSE ACETATE" />
<xs:enumeration value= "FORMVAR" />
<xs:enumeration value= "GOLD" />
<xs:enumeration value= "GRAPHENE" />
<xs:enumeration value= "GRAPHENE OXIDE" />
<xs:enumeration value= "PARLODION" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
CONTINUOUS
enumeration
HOLEY
enumeration
HOLEY ARRAY
enumeration
LACEY
Source
<xs:element name= "film_topology" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "CONTINUOUS" />
<xs:enumeration value= "HOLEY" />
<xs:enumeration value= "HOLEY ARRAY" />
<xs:enumeration value= "LACEY" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
AIR
enumeration
AMYLAMINE
enumeration
NITROGEN
enumeration
OTHER
Source
<xs:element name= "atmosphere" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "AIR" />
<xs:enumeration value= "AMYLAMINE" />
<xs:enumeration value= "NITROGEN" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type vitrification_type
Properties
content
complex
minOccurs
0
Model
cryogen_name ,
chamber_humidity{0,1} ,
chamber_temperature{0,1} ,
instrument{0,1} ,
details{0,1} ,
timed_resolved_state{0,1} ,
method{0,1}
Children chamber_humidity chamber_temperature cryogen_name details instrument method timed_resolved_state
Instance
<vitrification >
<cryogen_name > {1,1} </cryogen_name>
<chamber_humidity units= "percentage" > {0,1} </chamber_humidity>
<chamber_temperature units= "K" > {0,1} </chamber_temperature>
<instrument > {0,1} </instrument>
<details > {0,1} </details>
<timed_resolved_state > {0,1} </timed_resolved_state>
<method > {0,1} </method>
</vitrification>
Source
<xs:element name= "vitrification" type= "vitrification_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
ETHANE
enumeration
ETHANE-PROPANE
enumeration
FREON 12
enumeration
FREON 22
enumeration
HELIUM
enumeration
METHANE
enumeration
NITROGEN
enumeration
OTHER
enumeration
PROPANE
Source
<xs:element name= "cryogen_name" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "ETHANE" />
<xs:enumeration value= "ETHANE-PROPANE" />
<xs:enumeration value= "FREON 12" />
<xs:enumeration value= "FREON 22" />
<xs:enumeration value= "HELIUM" />
<xs:enumeration value= "METHANE" />
<xs:enumeration value= "NITROGEN" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PROPANE" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
EMS-002 RAPID IMMERSION FREEZER
enumeration
FEI VITROBOT MARK I
enumeration
FEI VITROBOT MARK II
enumeration
FEI VITROBOT MARK III
enumeration
FEI VITROBOT MARK IV
enumeration
GATAN CRYOPLUNGE 3
enumeration
HOMEMADE PLUNGER
enumeration
LEICA EM CPC
enumeration
LEICA EM GP
enumeration
LEICA KF80
enumeration
LEICA PLUNGER
enumeration
REICHERT-JUNG PLUNGER
enumeration
SPOTITON
enumeration
OTHER
Source
<xs:element name= "instrument" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EMS-002 RAPID IMMERSION FREEZER" />
<xs:enumeration value= "FEI VITROBOT MARK I" />
<xs:enumeration value= "FEI VITROBOT MARK II" />
<xs:enumeration value= "FEI VITROBOT MARK III" />
<xs:enumeration value= "FEI VITROBOT MARK IV" />
<xs:enumeration value= "GATAN CRYOPLUNGE 3" />
<xs:enumeration value= "HOMEMADE PLUNGER" />
<xs:enumeration value= "LEICA EM CPC" />
<xs:enumeration value= "LEICA EM GP" />
<xs:enumeration value= "LEICA KF80" />
<xs:enumeration value= "LEICA PLUNGER" />
<xs:enumeration value= "REICHERT-JUNG PLUNGER" />
<xs:enumeration value= "SPOTITON" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
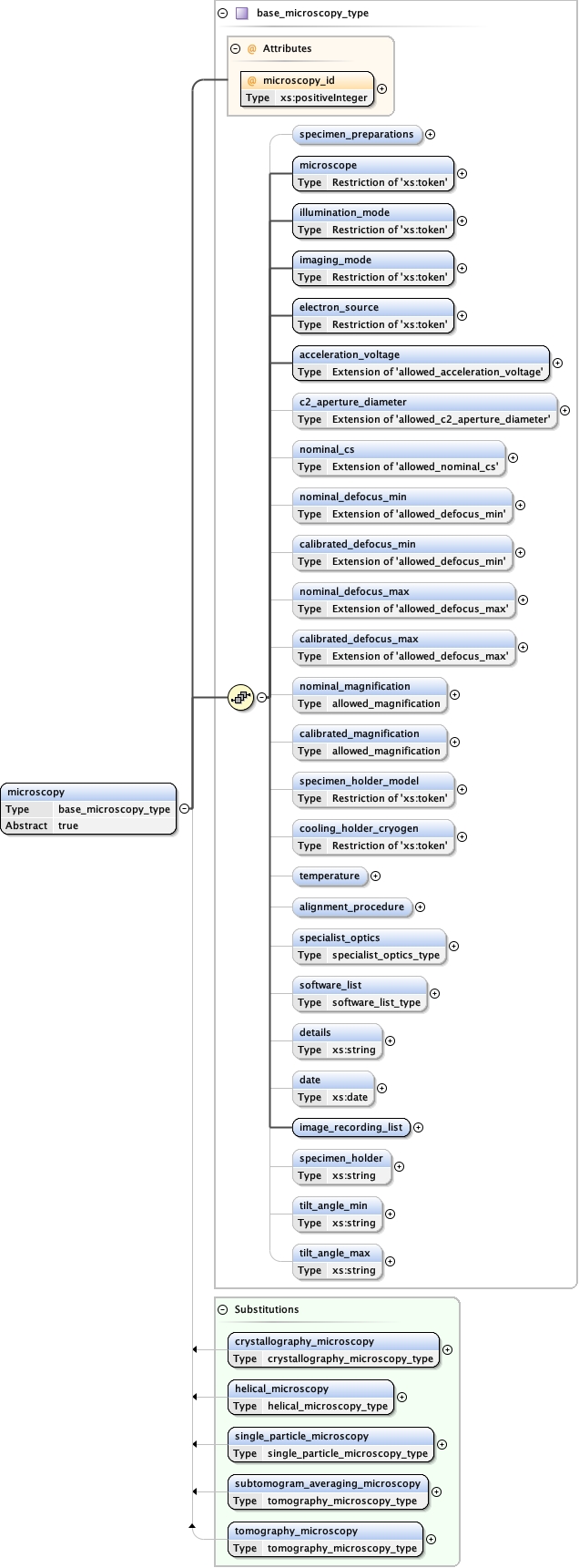
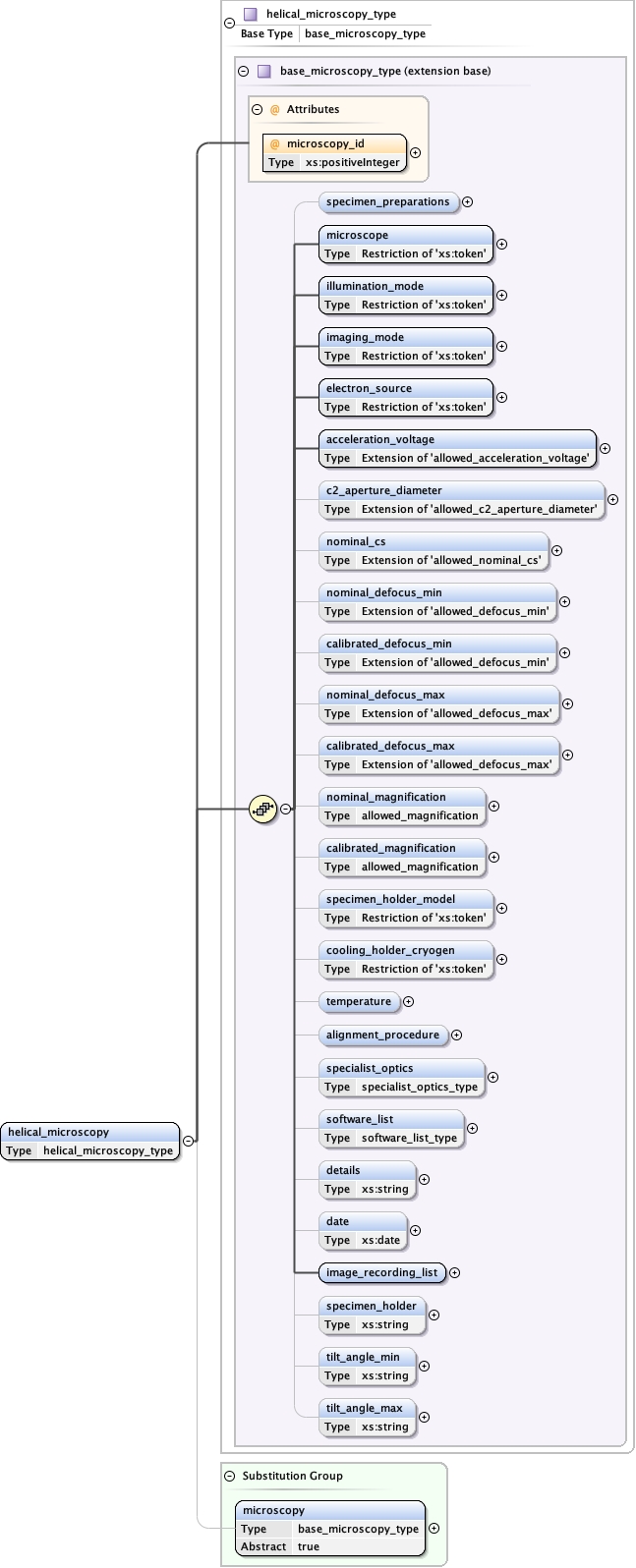
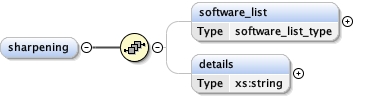
Element microscopy
Namespace No namespace
Diagram
Type base_microscopy_type
Properties
content
complex
abstract
true
Substitution Group
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
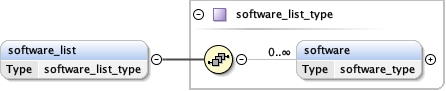
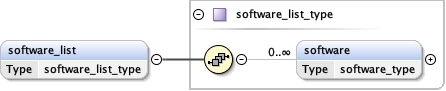
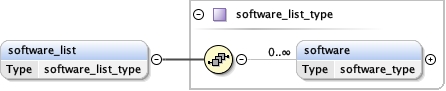
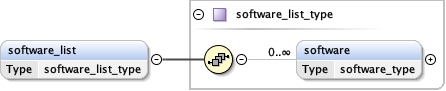
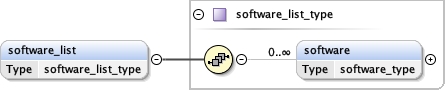
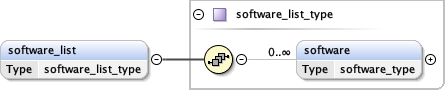
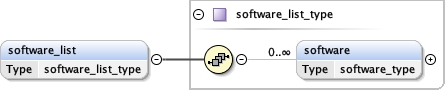
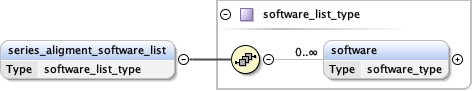
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
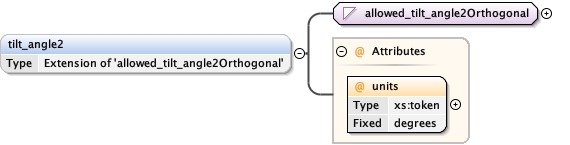
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Instance
<microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
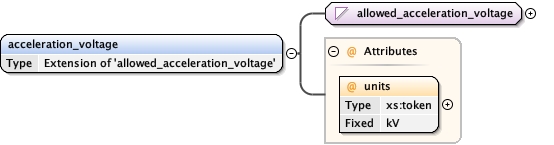
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
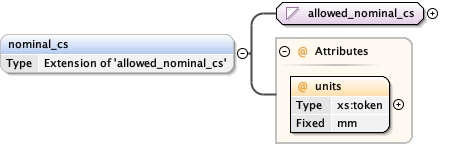
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
</microscopy>
Attributes
Source
<xs:element name= "microscopy" type= "base_microscopy_type" abstract= "true" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
FEI MORGAGNI
enumeration
FEI POLARA 300
enumeration
FEI TALOS ARCTICA
enumeration
FEI TECNAI 10
enumeration
FEI TECNAI 12
enumeration
FEI TECNAI 20
enumeration
FEI TECNAI ARCTICA
enumeration
FEI TECNAI F20
enumeration
FEI TECNAI F30
enumeration
FEI TECNAI SPHERA
enumeration
FEI TECNAI SPIRIT
enumeration
FEI TITAN
enumeration
FEI TITAN KRIOS
enumeration
FEI/PHILIPS CM10
enumeration
FEI/PHILIPS CM12
enumeration
FEI/PHILIPS CM120T
enumeration
FEI/PHILIPS CM200FEG
enumeration
FEI/PHILIPS CM200FEG/SOPHIE
enumeration
FEI/PHILIPS CM200FEG/ST
enumeration
FEI/PHILIPS CM200FEG/UT
enumeration
FEI/PHILIPS CM200T
enumeration
FEI/PHILIPS CM300FEG/HE
enumeration
FEI/PHILIPS CM300FEG/ST
enumeration
FEI/PHILIPS CM300FEG/T
enumeration
FEI/PHILIPS EM400
enumeration
FEI/PHILIPS EM420
enumeration
HITACHI EF2000
enumeration
HITACHI H-9500SD
enumeration
HITACHI H3000 UHVEM
enumeration
HITACHI H7600
enumeration
HITACHI HF2000
enumeration
HITACHI HF3000
enumeration
JEOL 100CX
enumeration
JEOL 1000EES
enumeration
JEOL 1010
enumeration
JEOL 1200
enumeration
JEOL 1200EX
enumeration
JEOL 1200EXII
enumeration
JEOL 1230
enumeration
JEOL 1400
enumeration
JEOL 2000EX
enumeration
JEOL 2000EXII
enumeration
JEOL 2010
enumeration
JEOL 2010F
enumeration
JEOL 2010HC
enumeration
JEOL 2010HT
enumeration
JEOL 2010UHR
enumeration
JEOL 2011
enumeration
JEOL 2100
enumeration
JEOL 2100F
enumeration
JEOL 2200FS
enumeration
JEOL 2200FSC
enumeration
JEOL 3000SFF
enumeration
JEOL 3100FEF
enumeration
JEOL 3100FFC
enumeration
JEOL 3200FS
enumeration
JEOL 3200FSC
enumeration
JEOL 4000
enumeration
JEOL 4000EX
enumeration
JEOL CRYO ARM 200
enumeration
JEOL CRYO ARM 300
enumeration
JEOL KYOTO-3000SFF
enumeration
TFS GLACIOS
enumeration
TFS KRIOS
enumeration
TFS TALOS
enumeration
TFS TALOS L120C
enumeration
TFS TALOS F200C
enumeration
ZEISS LEO912
enumeration
ZEISS LIBRA120PLUS
Source
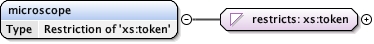
<xs:element name= "microscope" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "FEI MORGAGNI" />
<xs:enumeration value= "FEI POLARA 300" />
<xs:enumeration value= "FEI TALOS ARCTICA" />
<xs:enumeration value= "FEI TECNAI 10" />
<xs:enumeration value= "FEI TECNAI 12" />
<xs:enumeration value= "FEI TECNAI 20" />
<xs:enumeration value= "FEI TECNAI ARCTICA" />
<xs:enumeration value= "FEI TECNAI F20" />
<xs:enumeration value= "FEI TECNAI F30" />
<xs:enumeration value= "FEI TECNAI SPHERA" />
<xs:enumeration value= "FEI TECNAI SPIRIT" />
<xs:enumeration value= "FEI TITAN" />
<xs:enumeration value= "FEI TITAN KRIOS" />
<xs:enumeration value= "FEI/PHILIPS CM10" />
<xs:enumeration value= "FEI/PHILIPS CM12" />
<xs:enumeration value= "FEI/PHILIPS CM120T" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/SOPHIE" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/ST" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/UT" />
<xs:enumeration value= "FEI/PHILIPS CM200T" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/HE" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/ST" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/T" />
<xs:enumeration value= "FEI/PHILIPS EM400" />
<xs:enumeration value= "FEI/PHILIPS EM420" />
<xs:enumeration value= "HITACHI EF2000" />
<xs:enumeration value= "HITACHI H-9500SD" />
<xs:enumeration value= "HITACHI H3000 UHVEM" />
<xs:enumeration value= "HITACHI H7600" />
<xs:enumeration value= "HITACHI HF2000" />
<xs:enumeration value= "HITACHI HF3000" />
<xs:enumeration value= "JEOL 100CX" />
<xs:enumeration value= "JEOL 1000EES" />
<xs:enumeration value= "JEOL 1010" />
<xs:enumeration value= "JEOL 1200" />
<xs:enumeration value= "JEOL 1200EX" />
<xs:enumeration value= "JEOL 1200EXII" />
<xs:enumeration value= "JEOL 1230" />
<xs:enumeration value= "JEOL 1400" />
<xs:enumeration value= "JEOL 2000EX" />
<xs:enumeration value= "JEOL 2000EXII" />
<xs:enumeration value= "JEOL 2010" />
<xs:enumeration value= "JEOL 2010F" />
<xs:enumeration value= "JEOL 2010HC" />
<xs:enumeration value= "JEOL 2010HT" />
<xs:enumeration value= "JEOL 2010UHR" />
<xs:enumeration value= "JEOL 2011" />
<xs:enumeration value= "JEOL 2100" />
<xs:enumeration value= "JEOL 2100F" />
<xs:enumeration value= "JEOL 2200FS" />
<xs:enumeration value= "JEOL 2200FSC" />
<xs:enumeration value= "JEOL 3000SFF" />
<xs:enumeration value= "JEOL 3100FEF" />
<xs:enumeration value= "JEOL 3100FFC" />
<xs:enumeration value= "JEOL 3200FS" />
<xs:enumeration value= "JEOL 3200FSC" />
<xs:enumeration value= "JEOL 4000" />
<xs:enumeration value= "JEOL 4000EX" />
<xs:enumeration value= "JEOL CRYO ARM 200" />
<xs:enumeration value= "JEOL CRYO ARM 300" />
<xs:enumeration value= "JEOL KYOTO-3000SFF" />
<xs:enumeration value= "TFS GLACIOS" />
<xs:enumeration value= "TFS KRIOS" />
<xs:enumeration value= "TFS TALOS" />
<xs:enumeration value= "TFS TALOS L120C" />
<xs:enumeration value= "TFS TALOS F200C" />
<xs:enumeration value= "ZEISS LEO912" />
<xs:enumeration value= "ZEISS LIBRA120PLUS" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
BRIGHT FIELD
enumeration
DARK FIELD
enumeration
DIFFRACTION
enumeration
OTHER
Source
<xs:element name= "imaging_mode" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "BRIGHT FIELD" />
<xs:enumeration value= "DARK FIELD" />
<xs:enumeration value= "DIFFRACTION" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
TUNGSTEN HAIRPIN
enumeration
LAB6
enumeration
OTHER
enumeration
FIELD EMISSION GUN
Source
<xs:element name= "electron_source" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "TUNGSTEN HAIRPIN" />
<xs:enumeration value= "LAB6" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "FIELD EMISSION GUN" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
FISCHIONE 2550
enumeration
FEI TITAN KRIOS AUTOGRID HOLDER
enumeration
GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER
enumeration
GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER
enumeration
GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER
enumeration
GATAN 915 DOUBLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER
enumeration
GATAN CHDT 3504 DOUBLE TILT HIGH RESOLUTION NITROGEN COOLING HOLDER
enumeration
GATAN CT3500 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER
enumeration
GATAN CT3500TR SINGLE TILT ROTATION LIQUID NITROGEN CRYO TRANSFER HOLDER
enumeration
GATAN ELSA 698 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER
enumeration
GATAN HC 3500 SINGLE TILT HEATING/NITROGEN COOLING HOLDER
enumeration
GATAN HCHDT 3010 DOUBLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER
enumeration
GATAN HCHST 3008 SINGLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER
enumeration
GATAN HELIUM
enumeration
GATAN LIQUID NITROGEN
enumeration
GATAN UHRST 3500 SINGLE TILT ULTRA HIGH RESOLUTION NITROGEN COOLING HOLDER
enumeration
GATAN ULTDT ULTRA LOW TEMPERATURE DOUBLE TILT HELIUM COOLING HOLDER
enumeration
GATAN ULTST ULTRA LOW TEMPERATURE SINGLE TILT HELIUM COOLING HOLDER
enumeration
HOME BUILD
enumeration
JEOL
enumeration
JEOL 3200FSC CRYOHOLDER
enumeration
OTHER
enumeration
PHILIPS ROTATION HOLDER
enumeration
SIDE ENTRY, EUCENTRIC
enumeration
JEOL CRYOSPECPORTER
Source
<xs:element name= "specimen_holder_model" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "FISCHIONE 2550" />
<xs:enumeration value= "FEI TITAN KRIOS AUTOGRID HOLDER" />
<xs:enumeration value= "GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER" />
<xs:enumeration value= "GATAN 915 DOUBLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN CHDT 3504 DOUBLE TILT HIGH RESOLUTION NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN CT3500 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN CT3500TR SINGLE TILT ROTATION LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN ELSA 698 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN HC 3500 SINGLE TILT HEATING/NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN HCHDT 3010 DOUBLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN HCHST 3008 SINGLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN HELIUM" />
<xs:enumeration value= "GATAN LIQUID NITROGEN" />
<xs:enumeration value= "GATAN UHRST 3500 SINGLE TILT ULTRA HIGH RESOLUTION NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN ULTDT ULTRA LOW TEMPERATURE DOUBLE TILT HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN ULTST ULTRA LOW TEMPERATURE SINGLE TILT HELIUM COOLING HOLDER" />
<xs:enumeration value= "HOME BUILD" />
<xs:enumeration value= "JEOL" />
<xs:enumeration value= "JEOL 3200FSC CRYOHOLDER" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PHILIPS ROTATION HOLDER" />
<xs:enumeration value= "SIDE ENTRY, EUCENTRIC" />
<xs:enumeration value= "JEOL CRYOSPECPORTER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Properties
content
complex
minOccurs
0
Model
Children basic coma_free legacy none other zemlin_tableau
Instance
<alignment_procedure >
<none > {1,1} </none>
<basic > {1,1} </basic>
<zemlin_tableau > {1,1} </zemlin_tableau>
<coma_free > {1,1} </coma_free>
<other > {1,1} </other>
<legacy > {1,1} </legacy>
</alignment_procedure>
Source
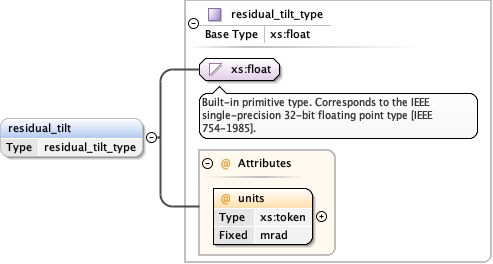
<xs:element name= "alignment_procedure" minOccurs= "0" >
<xs:complexType >
<xs:choice >
<xs:element name= "none" >
<xs:complexType />
</xs:element>
<xs:element name= "basic" >
<xs:complexType >
<xs:sequence >
<xs:element name= "residual_tilt" type= "residual_tilt_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "zemlin_tableau" >
<xs:complexType />
</xs:element>
<xs:element name= "coma_free" >
<xs:complexType >
<xs:sequence >
<xs:element name= "residual_tilt" type= "residual_tilt_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "other" >
<xs:complexType />
</xs:element>
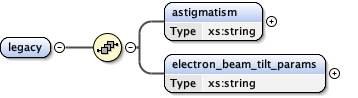
<xs:element name= "legacy" >
<xs:complexType >
<xs:sequence >
<xs:element name= "astigmatism" type= "xs:string" />
<xs:element name= "electron_beam_tilt_params" type= "xs:string" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:choice>
</xs:complexType>
</xs:element>
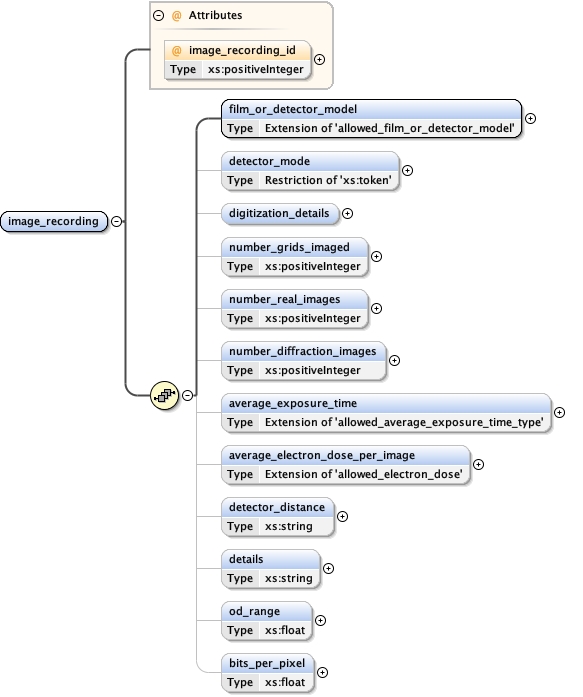
Namespace No namespace
Diagram
Properties
Model
film_or_detector_model ,
detector_mode{0,1} ,
digitization_details{0,1} ,
number_grids_imaged{0,1} ,
number_real_images{0,1} ,
number_diffraction_images{0,1} ,
average_exposure_time{0,1} ,
average_electron_dose_per_image{0,1} ,
detector_distance{0,1} ,
details{0,1} ,
od_range{0,1} ,
bits_per_pixel{0,1}
Children average_electron_dose_per_image average_exposure_time bits_per_pixel details detector_distance detector_mode digitization_details film_or_detector_model number_diffraction_images number_grids_imaged number_real_images od_range
Instance
<image_recording image_recording_id= "" >
<film_or_detector_model category= "" > {1,1} </film_or_detector_model>
<detector_mode > {0,1} </detector_mode>
<digitization_details > {0,1} </digitization_details>
<number_grids_imaged > {0,1} </number_grids_imaged>
<number_real_images > {0,1} </number_real_images>
<number_diffraction_images > {0,1} </number_diffraction_images>
<average_exposure_time units= "s" > {0,1} </average_exposure_time>
<average_electron_dose_per_image units= "e/Å^2" > {0,1} </average_electron_dose_per_image>
<detector_distance > {0,1} </detector_distance>
<details > {0,1} </details>
<od_range > {0,1} </od_range>
<bits_per_pixel > {0,1} </bits_per_pixel>
</image_recording>
Attributes
Source
<xs:element name= "image_recording" >
<xs:complexType >
<xs:sequence >
<xs:element name= "film_or_detector_model" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_film_or_detector_model" >
<xs:attribute name= "category" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "CCD" />
<xs:enumeration value= "CMOS" />
<xs:enumeration value= "DIRECT ELECTRON DETECTOR" />
<xs:enumeration value= "STORAGE PHOSPOR (IMAGE PLATES)" />
<xs:enumeration value= "FILM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "detector_mode" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "COUNTING" />
<xs:enumeration value= "INTEGRATING" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "SUPER-RESOLUTION" />
</xs:restriction>
</xs:simpleType>
</xs:element>
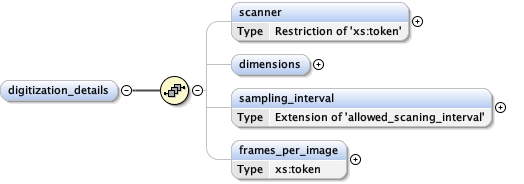
<xs:element name= "digitization_details" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "scanner" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EIKONIX IEEE 488" />
<xs:enumeration value= "EMIL 10" />
<xs:enumeration value= "IMACON" />
<xs:enumeration value= "NIKON COOLSCAN" />
<xs:enumeration value= "NIKON SUPER COOLSCAN 9000" />
<xs:enumeration value= "OPTRONICS" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PATCHWORK DENSITOMETER" />
<xs:enumeration value= "PERKIN ELMER" />
<xs:enumeration value= "PRIMESCAN" />
<xs:enumeration value= "TEMSCAN" />
<xs:enumeration value= "ZEISS SCAI" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "dimensions" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "width" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:positiveInteger" >
<xs:attribute name= "units" type= "xs:token" fixed= "pixel" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "height" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:positiveInteger" >
<xs:attribute name= "units" type= "xs:token" fixed= "pixel" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "sampling_interval" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_scaning_interval" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "frames_per_image" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "number_grids_imaged" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "number_real_images" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "number_diffraction_images" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "average_exposure_time" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_average_exposure_time_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "s" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "average_electron_dose_per_image" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_electron_dose" >
<xs:attribute name= "units" type= "xs:token" fixed= "e/Å^2" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "detector_distance" type= "xs:string" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "od_range" type= "xs:float" minOccurs= "0" />
<xs:element name= "bits_per_pixel" type= "xs:float" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "image_recording_id" type= "xs:positiveInteger" />
</xs:complexType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
COUNTING
enumeration
INTEGRATING
enumeration
OTHER
enumeration
SUPER-RESOLUTION
Source
<xs:element name= "detector_mode" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "COUNTING" />
<xs:enumeration value= "INTEGRATING" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "SUPER-RESOLUTION" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
EIKONIX IEEE 488
enumeration
EMIL 10
enumeration
IMACON
enumeration
NIKON COOLSCAN
enumeration
NIKON SUPER COOLSCAN 9000
enumeration
OPTRONICS
enumeration
OTHER
enumeration
PATCHWORK DENSITOMETER
enumeration
PERKIN ELMER
enumeration
PRIMESCAN
enumeration
TEMSCAN
enumeration
ZEISS SCAI
Source
<xs:element name= "scanner" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EIKONIX IEEE 488" />
<xs:enumeration value= "EMIL 10" />
<xs:enumeration value= "IMACON" />
<xs:enumeration value= "NIKON COOLSCAN" />
<xs:enumeration value= "NIKON SUPER COOLSCAN 9000" />
<xs:enumeration value= "OPTRONICS" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PATCHWORK DENSITOMETER" />
<xs:enumeration value= "PERKIN ELMER" />
<xs:enumeration value= "PRIMESCAN" />
<xs:enumeration value= "TEMSCAN" />
<xs:enumeration value= "ZEISS SCAI" />
</xs:restriction>
</xs:simpleType>
</xs:element>
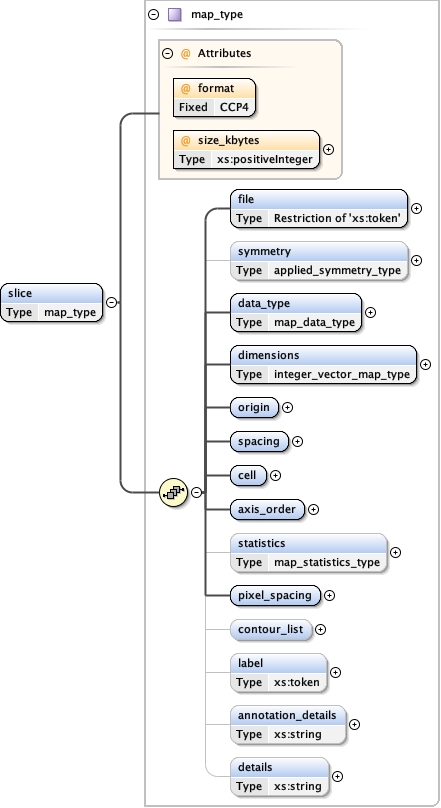
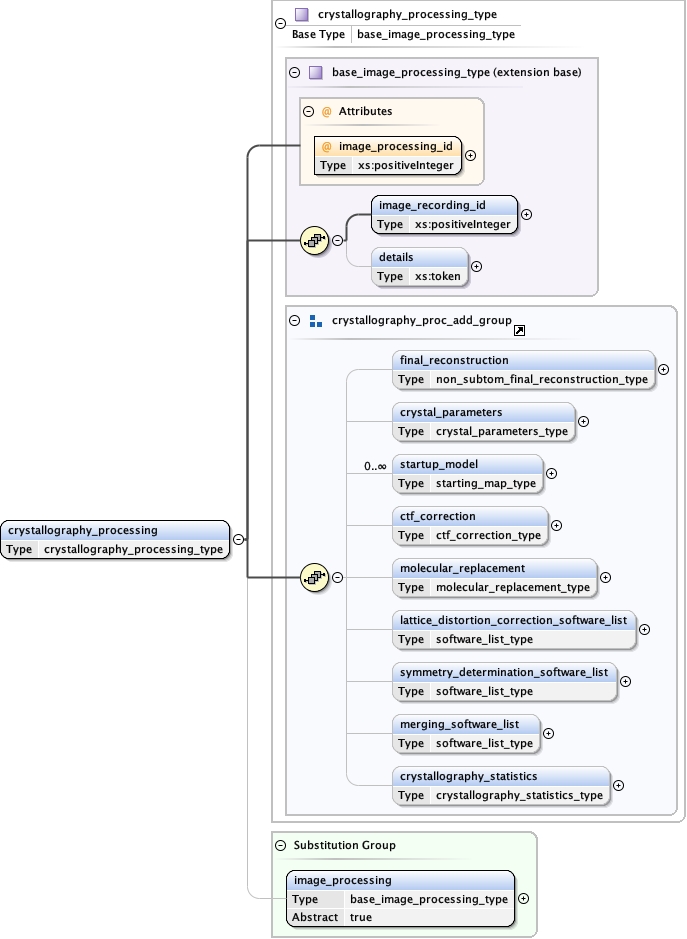
Element image_processing
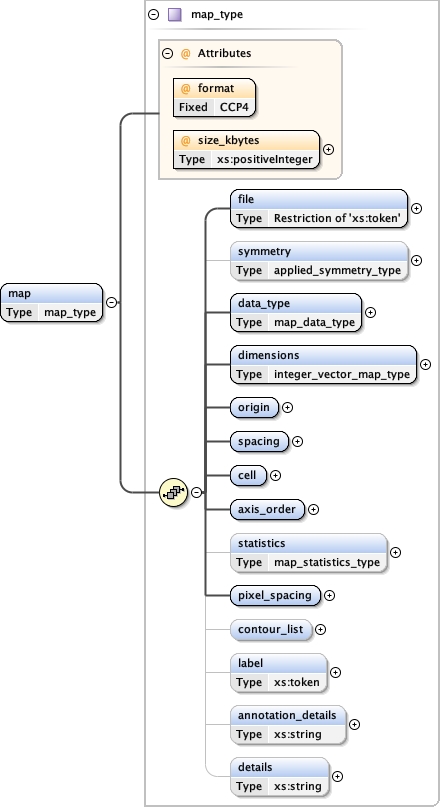
Namespace No namespace
Diagram
Type map_type
Properties
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Instance
<map format= "CCP4" size_kbytes= "" >
<file > {1,1} </file>
<symmetry > {0,1} </symmetry>
<data_type > {1,1} </data_type>
<dimensions > {1,1} </dimensions>
<origin > {1,1} </origin>
<spacing > {1,1} </spacing>
<cell > {1,1} </cell>
<axis_order > {1,1} </axis_order>
<statistics > {0,1} </statistics>
<pixel_spacing > {1,1} </pixel_spacing>
<contour_list > {0,1} </contour_list>
<label > {0,1} </label>
<annotation_details > {0,1} </annotation_details>
<details > {0,1} </details>
</map>
Attributes
Source
<xs:element name= "map" type= "map_type" />
Namespace No namespace
Diagram
Properties
Model
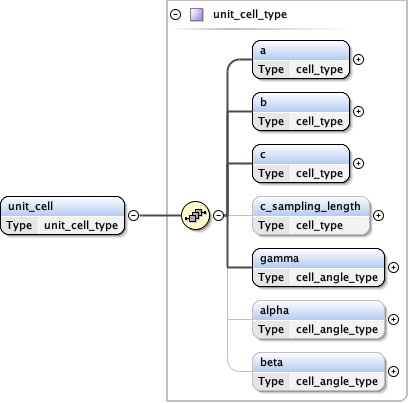
Children a alpha b beta c gamma
Instance
<cell >
<a units= "Å" > {1,1} </a>
<b units= "Å" > {1,1} </b>
<c units= "Å" > {1,1} </c>
<alpha units= "deg" > {1,1} </alpha>
<beta units= "deg" > {1,1} </beta>
<gamma units= "deg" > {1,1} </gamma>
</cell>
Source
<xs:element name= "cell" >
<xs:complexType >
<xs:sequence >
<xs:element name= "a" type= "cell_type" />
<xs:element name= "b" type= "cell_type" />
<xs:element name= "c" type= "cell_type" />
<xs:element name= "alpha" type= "cell_angle_type" />
<xs:element name= "beta" type= "cell_angle_type" />
<xs:element name= "gamma" type= "cell_angle_type" />
</xs:sequence>
</xs:complexType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
EMDB
enumeration
AUTHOR
enumeration
SOFTWARE
enumeration
ANNOTATOR
Source
<xs:element name= "source" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EMDB" />
<xs:enumeration value= "AUTHOR" />
<xs:enumeration value= "SOFTWARE" />
<xs:enumeration value= "ANNOTATOR" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type interpretation_type
Properties
content
complex
minOccurs
0
Model
modelling_list{0,1} ,
figure_list{0,1} ,
segmentation_list{0,1} ,
slices_list{0,1} ,
additional_map_list{0,1} ,
half_map_list{0,1}
Children additional_map_list figure_list half_map_list modelling_list segmentation_list slices_list
Instance
<interpretation >
<modelling_list > {0,1} </modelling_list>
<figure_list > {0,1} </figure_list>
<segmentation_list > {0,1} </segmentation_list>
<slices_list > {0,1} </slices_list>
<additional_map_list > {0,1} </additional_map_list>
<half_map_list > {0,1} </half_map_list>
</interpretation>
Source
<xs:element name= "interpretation" type= "interpretation_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type modelling_type
Properties
content
complex
maxOccurs
unbounded
Model
initial_model* ,
final_model{0,1} ,
refinement_protocol{0,1} ,
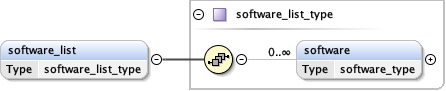
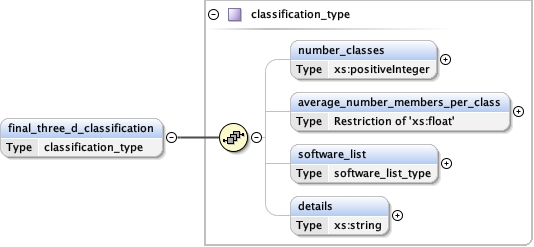
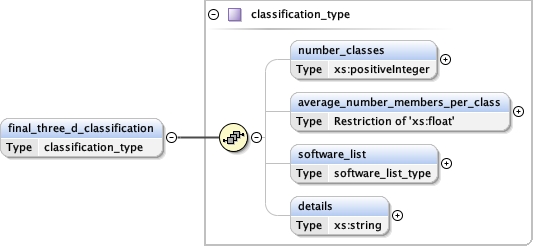
software_list{0,1} ,
details{0,1} ,
target_criteria{0,1} ,
refinement_space{0,1} ,
overall_bvalue{0,1}
Children details final_model initial_model overall_bvalue refinement_protocol refinement_space software_list target_criteria
Instance
<modelling >
<initial_model > {0,unbounded} </initial_model>
<final_model > {0,1} </final_model>
<refinement_protocol > {0,1} </refinement_protocol>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<target_criteria > {0,1} </target_criteria>
<refinement_space > {0,1} </refinement_space>
<overall_bvalue > {0,1} </overall_bvalue>
</modelling>
Source
<xs:element name= "modelling" type= "modelling_type" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type restriction of xs:string
Properties
content
simple
minOccurs
0
Facets
enumeration
AB INITIO MODEL
enumeration
BACKBONE TRACE
enumeration
FLEXIBLE FIT
enumeration
OTHER
enumeration
RIGID BODY FIT
Source
<xs:element name= "refinement_protocol" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "AB INITIO MODEL" />
<xs:enumeration value= "BACKBONE TRACE" />
<xs:enumeration value= "FLEXIBLE FIT" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "RIGID BODY FIT" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type map_type
Properties
content
complex
minOccurs
0
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Instance
<mask_details format= "CCP4" size_kbytes= "" >
<file > {1,1} </file>
<symmetry > {0,1} </symmetry>
<data_type > {1,1} </data_type>
<dimensions > {1,1} </dimensions>
<origin > {1,1} </origin>
<spacing > {1,1} </spacing>
<cell > {1,1} </cell>
<axis_order > {1,1} </axis_order>
<statistics > {0,1} </statistics>
<pixel_spacing > {1,1} </pixel_spacing>
<contour_list > {0,1} </contour_list>
<label > {0,1} </label>
<annotation_details > {0,1} </annotation_details>
<details > {0,1} </details>
</mask_details>
Attributes
Source
<xs:element name= "mask_details" type= "map_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type map_type
Properties
content
complex
maxOccurs
unbounded
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Instance
<slice format= "CCP4" size_kbytes= "" >
<file > {1,1} </file>
<symmetry > {0,1} </symmetry>
<data_type > {1,1} </data_type>
<dimensions > {1,1} </dimensions>
<origin > {1,1} </origin>
<spacing > {1,1} </spacing>
<cell > {1,1} </cell>
<axis_order > {1,1} </axis_order>
<statistics > {0,1} </statistics>
<pixel_spacing > {1,1} </pixel_spacing>
<contour_list > {0,1} </contour_list>
<label > {0,1} </label>
<annotation_details > {0,1} </annotation_details>
<details > {0,1} </details>
</slice>
Attributes
Source
<xs:element name= "slice" type= "map_type" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type map_type
Properties
content
complex
maxOccurs
unbounded
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Instance
<additional_map format= "CCP4" size_kbytes= "" >
<file > {1,1} </file>
<symmetry > {0,1} </symmetry>
<data_type > {1,1} </data_type>
<dimensions > {1,1} </dimensions>
<origin > {1,1} </origin>
<spacing > {1,1} </spacing>
<cell > {1,1} </cell>
<axis_order > {1,1} </axis_order>
<statistics > {0,1} </statistics>
<pixel_spacing > {1,1} </pixel_spacing>
<contour_list > {0,1} </contour_list>
<label > {0,1} </label>
<annotation_details > {0,1} </annotation_details>
<details > {0,1} </details>
</additional_map>
Attributes
Source
<xs:element name= "additional_map" type= "map_type" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type map_type
Properties
content
complex
maxOccurs
unbounded
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Instance
<half_map format= "CCP4" size_kbytes= "" >
<file > {1,1} </file>
<symmetry > {0,1} </symmetry>
<data_type > {1,1} </data_type>
<dimensions > {1,1} </dimensions>
<origin > {1,1} </origin>
<spacing > {1,1} </spacing>
<cell > {1,1} </cell>
<axis_order > {1,1} </axis_order>
<statistics > {0,1} </statistics>
<pixel_spacing > {1,1} </pixel_spacing>
<contour_list > {0,1} </contour_list>
<label > {0,1} </label>
<annotation_details > {0,1} </annotation_details>
<details > {0,1} </details>
</half_map>
Attributes
Source
<xs:element name= "half_map" type= "map_type" maxOccurs= "unbounded" />
Element validation_method
Element journal_citation
Namespace No namespace
Diagram
Properties
Substitution Group Affiliation
Model
author+ ,
title ,
journal{0,1} ,
journal_abbreviation ,
country{0,1} ,
issue{0,1} ,
volume{0,1} ,
first_page{0,1} ,
last_page{0,1} ,
year{0,1} ,
language{0,1} ,
external_references* ,
details{0,1}
Children author country details external_references first_page issue journal journal_abbreviation language last_page title volume year
Instance
<journal_citation published= "" >
<author order= "" > {1,unbounded} </author>
<title > {1,1} </title>
<journal > {0,1} </journal>
<journal_abbreviation > {1,1} </journal_abbreviation>
<country > {0,1} </country>
<issue > {0,1} </issue>
<volume > {0,1} </volume>
<first_page > {0,1} </first_page>
<last_page > {0,1} </last_page>
<year > {0,1} </year>
<language > {0,1} </language>
<external_references type= "" > {0,unbounded} </external_references>
<details > {0,1} </details>
</journal_citation>
Attributes
Source
<xs:element name= "journal_citation" substitutionGroup= "citation_type" >
<xs:complexType >
<xs:sequence >
<xs:element name= "author" type= "author_order_type" maxOccurs= "unbounded" />
<xs:element name= "title" type= "xs:token" />
<xs:element name= "journal" type= "xs:token" minOccurs= "0" />
<xs:element name= "journal_abbreviation" type= "xs:token" />
<xs:element name= "country" type= "xs:token" minOccurs= "0" />
<xs:element name= "issue" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "volume" type= "xs:string" nillable= "true" minOccurs= "0" />
<xs:element name= "first_page" type= "page_type" nillable= "false" minOccurs= "0" />
<xs:element name= "last_page" type= "page_type" minOccurs= "0" />
<xs:element name= "year" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:gYear" >
<xs:minInclusive value= "1900" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "language" type= "xs:language" minOccurs= "0" />
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PUBMED" />
<xs:enumeration value= "DOI" />
<xs:enumeration value= "ISBN" />
<xs:enumeration value= "ISSN" />
<xs:enumeration value= "CAS" />
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CSD" />
<xs:enumeration value= "MEDLINE" />
<xs:enumeration value= "ASTM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "published" type= "xs:boolean" use= "required" />
</xs:complexType>
</xs:element>
Element non_journal_citation
Namespace No namespace
Diagram
Properties
Substitution Group Affiliation
Model
author+ ,
editor* ,
title ,
thesis_title{0,1} ,
chapter_title{0,1} ,
volume{0,1} ,
publisher{0,1} ,
publisher_location{0,1} ,
first_page{0,1} ,
last_page{0,1} ,
year ,
language{0,1} ,
external_references* ,
details{0,1}
Children author chapter_title details editor external_references first_page language last_page publisher publisher_location thesis_title title volume year
Instance
<non_journal_citation published= "" >
<author order= "" > {1,unbounded} </author>
<editor order= "" > {0,unbounded} </editor>
<title > {1,1} </title>
<thesis_title > {0,1} </thesis_title>
<chapter_title > {0,1} </chapter_title>
<volume > {0,1} </volume>
<publisher > {0,1} </publisher>
<publisher_location > {0,1} </publisher_location>
<first_page > {0,1} </first_page>
<last_page > {0,1} </last_page>
<year > {1,1} </year>
<language > {0,1} </language>
<external_references type= "" > {0,unbounded} </external_references>
<details > {0,1} </details>
</non_journal_citation>
Attributes
Source
<xs:element name= "non_journal_citation" substitutionGroup= "citation_type" >
<xs:complexType >
<xs:sequence >
<xs:element name= "author" type= "author_order_type" maxOccurs= "unbounded" />
<xs:element name= "editor" type= "author_order_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "title" type= "xs:token" />
<xs:element name= "thesis_title" type= "xs:token" minOccurs= "0" />
<xs:element name= "chapter_title" type= "xs:token" minOccurs= "0" />
<xs:element name= "volume" type= "xs:string" minOccurs= "0" />
<xs:element name= "publisher" type= "xs:token" minOccurs= "0" />
<xs:element name= "publisher_location" type= "xs:token" minOccurs= "0" />
<xs:element name= "first_page" type= "page_type" minOccurs= "0" />
<xs:element name= "last_page" type= "page_type" minOccurs= "0" />
<xs:element name= "year" >
<xs:simpleType >
<xs:restriction base= "xs:gYear" >
<xs:minInclusive value= "1900" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "language" type= "xs:language" minOccurs= "0" />
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PUBMED" />
<xs:enumeration value= "DOI" />
<xs:enumeration value= "ISBN" />
<xs:enumeration value= "ISSN" />
<xs:enumeration value= "CAS" />
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CSD" />
<xs:enumeration value= "MEDLINE" />
<xs:enumeration value= "ASTM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "published" type= "xs:boolean" use= "required" />
</xs:complexType>
</xs:element>
Element cell_supramolecule
Namespace No namespace
Diagram
Type cell_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source*
Children category details external_references macromolecule_list name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag synthetic_source
Instance
<cell_supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<natural_source database= "" > {0,unbounded} </natural_source>
<synthetic_source database= "" > {0,unbounded} </synthetic_source>
</cell_supramolecule>
Attributes
Source
<xs:element name= "cell_supramolecule" substitutionGroup= "supramolecule" type= "cell_supramolecule_type" />
Namespace No namespace
Diagram
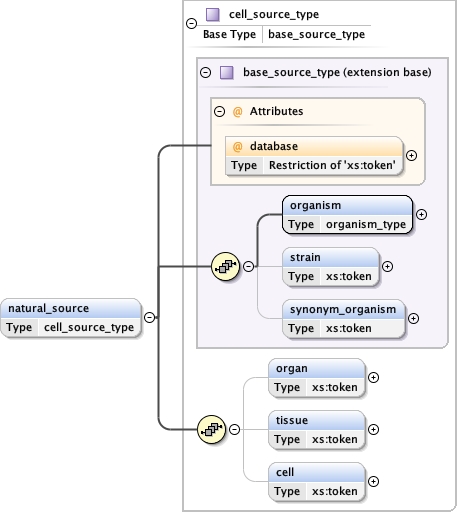
Type cell_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Instance
<natural_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
</natural_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "natural_source" type= "cell_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type cell_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "cell_source_type" minOccurs= "0" maxOccurs= "unbounded" />
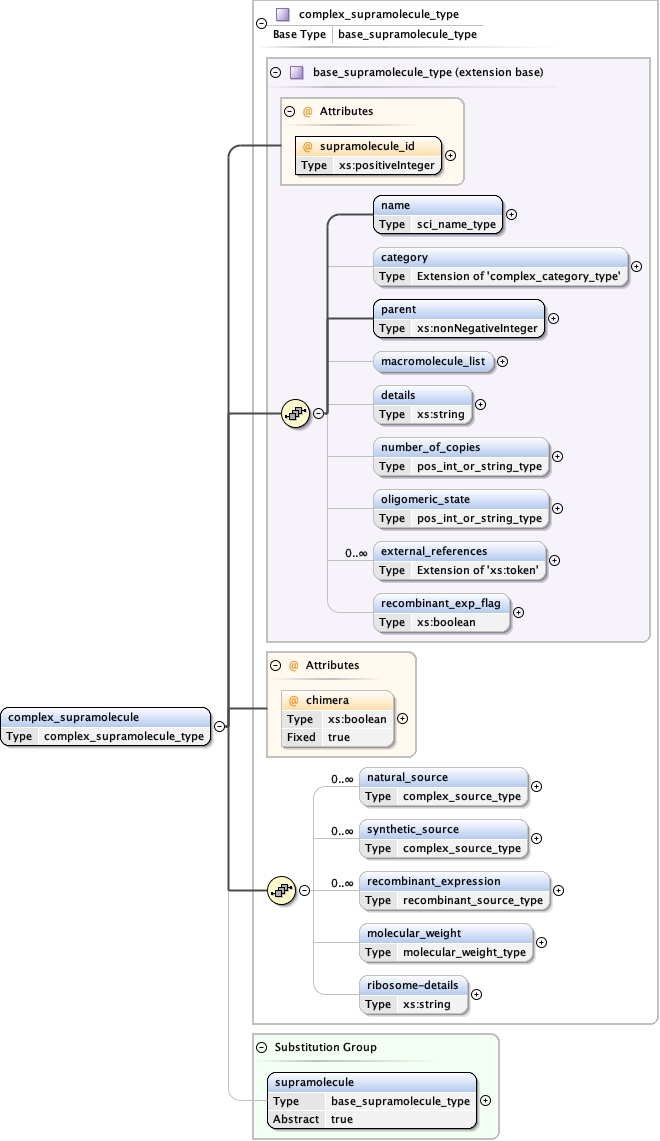
Element complex_supramolecule
Namespace No namespace
Diagram
Type complex_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
recombinant_expression* ,
molecular_weight{0,1} ,
ribosome-details{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag recombinant_expression ribosome-details synthetic_source
Instance
<complex_supramolecule chimera= "true" supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<natural_source database= "" > {0,unbounded} </natural_source>
<synthetic_source database= "" > {0,unbounded} </synthetic_source>
<recombinant_expression database= "" > {0,unbounded} </recombinant_expression>
<molecular_weight > {0,1} </molecular_weight>
<ribosome-details > {0,1} </ribosome-details>
</complex_supramolecule>
Attributes
Source
<xs:element name= "complex_supramolecule" substitutionGroup= "supramolecule" type= "complex_supramolecule_type" />
Namespace No namespace
Diagram
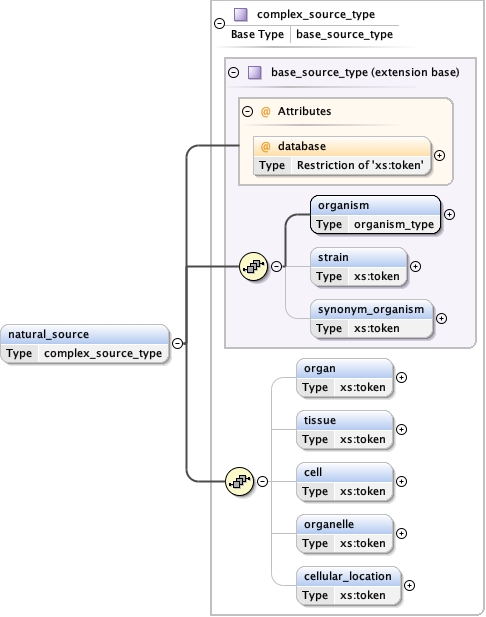
Type complex_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<natural_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</natural_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "natural_source" type= "complex_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type complex_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "complex_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Element organelle_or_cellular_component_supramolecule
Namespace No namespace
Diagram
Type organelle_or_cellular_component_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
molecular_weight{0,1} ,
recombinant_expression{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag recombinant_expression synthetic_source
Instance
<organelle_or_cellular_component_supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<natural_source database= "" > {0,unbounded} </natural_source>
<synthetic_source database= "" > {0,unbounded} </synthetic_source>
<molecular_weight > {0,1} </molecular_weight>
<recombinant_expression database= "" > {0,1} </recombinant_expression>
</organelle_or_cellular_component_supramolecule>
Attributes
Source
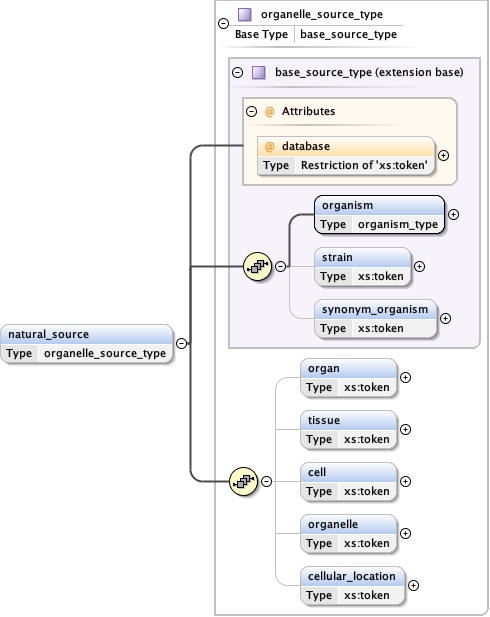
<xs:element name= "organelle_or_cellular_component_supramolecule" substitutionGroup= "supramolecule" type= "organelle_or_cellular_component_supramolecule_type" />
Namespace No namespace
Diagram
Type organelle_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<natural_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</natural_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "natural_source" type= "organelle_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type organelle_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "organelle_source_type" minOccurs= "0" maxOccurs= "unbounded" />
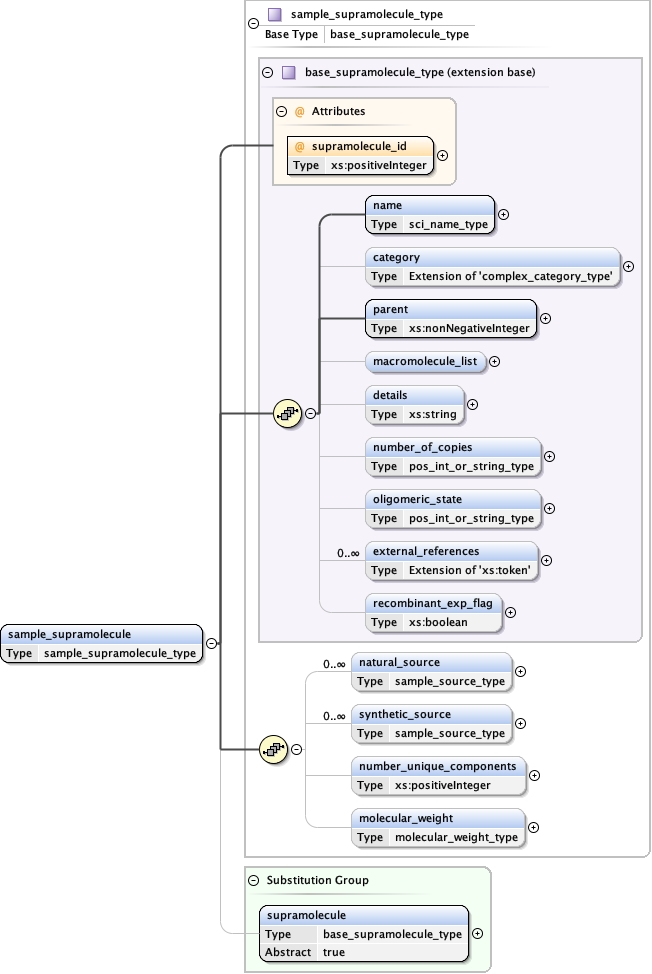
Element sample_supramolecule
Namespace No namespace
Diagram
Type sample_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
number_unique_components{0,1} ,
molecular_weight{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies number_unique_components oligomeric_state parent recombinant_exp_flag synthetic_source
Instance
<sample_supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<natural_source database= "" > {0,unbounded} </natural_source>
<synthetic_source database= "" > {0,unbounded} </synthetic_source>
<number_unique_components > {0,1} </number_unique_components>
<molecular_weight > {0,1} </molecular_weight>
</sample_supramolecule>
Attributes
Source
<xs:element name= "sample_supramolecule" substitutionGroup= "supramolecule" type= "sample_supramolecule_type" />
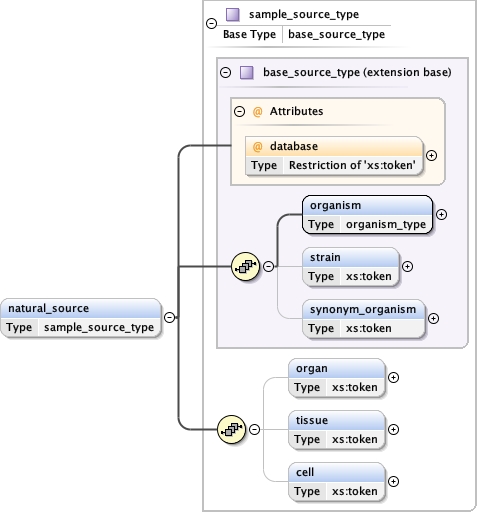
Namespace No namespace
Diagram
Type sample_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Instance
<natural_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
</natural_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "natural_source" type= "sample_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type sample_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "sample_source_type" minOccurs= "0" maxOccurs= "unbounded" />
Element tissue_supramolecule
Namespace No namespace
Diagram
Type tissue_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
sythetic_source*
Children category details external_references macromolecule_list name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag sythetic_source
Instance
<tissue_supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<natural_source database= "" > {0,unbounded} </natural_source>
<sythetic_source database= "" > {0,unbounded} </sythetic_source>
</tissue_supramolecule>
Attributes
Source
<xs:element name= "tissue_supramolecule" substitutionGroup= "supramolecule" type= "tissue_supramolecule_type" />
Element virus_supramolecule
Namespace No namespace
Diagram
Type virus_supramolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
sci_species_name{0,1} ,
sci_species_strain{0,1} ,
natural_host* ,
synthetic_host* ,
host_system{0,1} ,
molecular_weight{0,1} ,
virus_shell* ,
virus_type ,
virus_isolate ,
virus_enveloped ,
virus_empty ,
syn_species_name{0,1} ,
sci_species_serotype{0,1} ,
sci_species_serocomplex{0,1} ,
sci_species_subspecies{0,1}
Children category details external_references host_system macromolecule_list molecular_weight name natural_host number_of_copies oligomeric_state parent recombinant_exp_flag sci_species_name sci_species_serocomplex sci_species_serotype sci_species_strain sci_species_subspecies syn_species_name synthetic_host virus_empty virus_enveloped virus_isolate virus_shell virus_type
Instance
<virus_supramolecule supramolecule_id= "" >
<name synonym= "" > {1,1} </name>
<category type= "" > {0,1} </category>
<parent > {1,1} </parent>
<macromolecule_list > {0,1} </macromolecule_list>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<sci_species_name ncbi= "" > {0,1} </sci_species_name>
<sci_species_strain > {0,1} </sci_species_strain>
<natural_host database= "" > {0,unbounded} </natural_host>
<synthetic_host database= "" > {0,unbounded} </synthetic_host>
<host_system database= "" > {0,1} </host_system>
<molecular_weight > {0,1} </molecular_weight>
<virus_shell shell_id= "" > {0,unbounded} </virus_shell>
<virus_type > {1,1} </virus_type>
<virus_isolate > {1,1} </virus_isolate>
<virus_enveloped > {1,1} </virus_enveloped>
<virus_empty > {1,1} </virus_empty>
<syn_species_name > {0,1} </syn_species_name>
<sci_species_serotype > {0,1} </sci_species_serotype>
<sci_species_serocomplex > {0,1} </sci_species_serocomplex>
<sci_species_subspecies > {0,1} </sci_species_subspecies>
</virus_supramolecule>
Attributes
Source
<xs:element name= "virus_supramolecule" substitutionGroup= "supramolecule" type= "virus_supramolecule_type" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
PRION
enumeration
SATELLITE
enumeration
VIRION
enumeration
VIROID
enumeration
VIRUS-LIKE PARTICLE
Source
<xs:element name= "virus_type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PRION" />
<xs:enumeration value= "SATELLITE" />
<xs:enumeration value= "VIRION" />
<xs:enumeration value= "VIROID" />
<xs:enumeration value= "VIRUS-LIKE PARTICLE" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
OTHER
enumeration
SEROCOMPLEX
enumeration
SEROTYPE
enumeration
SPECIES
enumeration
STRAIN
enumeration
SUBSPECIES
Source
<xs:element name= "virus_isolate" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "SEROCOMPLEX" />
<xs:enumeration value= "SEROTYPE" />
<xs:enumeration value= "SPECIES" />
<xs:enumeration value= "STRAIN" />
<xs:enumeration value= "SUBSPECIES" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Element dna
Namespace No namespace
Diagram
Type dna_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence ,
classification{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1}
Children classification details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag sequence structure synthetic_flag synthetic_source
Instance
<dna chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<sequence > {1,1} </sequence>
<classification > {0,1} </classification>
<structure > {0,1} </structure>
<synthetic_flag > {0,1} </synthetic_flag>
<synthetic_source database= "" > {0,1} </synthetic_source>
</dna>
Attributes
Source
<xs:element name= "dna" type= "dna_macromolecule_type" substitutionGroup= "macromolecule" />
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
Element em_label
Namespace No namespace
Diagram
Type em_label_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
formula{0,1} ,
synthetic_source{0,1}
Children details formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag synthetic_source
Instance
<em_label chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<formula > {0,1} </formula>
<synthetic_source database= "" > {0,1} </synthetic_source>
</em_label>
Attributes
Source
<xs:element name= "em_label" type= "em_label_macromolecule_type" substitutionGroup= "macromolecule" />
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
Element ligand
Namespace No namespace
Diagram
Type ligand_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
formula{0,1} ,
external_references* ,
recombinant_expression{0,1}
Children details external_references formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression
Instance
<ligand chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<formula > {0,1} </formula>
<external_references type= "" > {0,unbounded} </external_references>
<recombinant_expression database= "" > {0,1} </recombinant_expression>
</ligand>
Attributes
Source
<xs:element name= "ligand" type= "ligand_macromolecule_type" substitutionGroup= "macromolecule" />
Element other_macromolecule
Namespace No namespace
Diagram
Type other_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence{0,1} ,
classification ,
recombinant_expression{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1}
Children classification details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression sequence structure synthetic_flag synthetic_source
Instance
<other_macromolecule chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<sequence > {0,1} </sequence>
<classification > {1,1} </classification>
<recombinant_expression database= "" > {0,1} </recombinant_expression>
<structure > {0,1} </structure>
<synthetic_flag > {0,1} </synthetic_flag>
<synthetic_source database= "" > {0,1} </synthetic_source>
</other_macromolecule>
Attributes
Source
<xs:element name= "other_macromolecule" type= "other_macromolecule_type" substitutionGroup= "macromolecule" />
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
Element protein_or_peptide
Namespace No namespace
Diagram
Type protein_or_peptide_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
recombinant_expression{0,1} ,
synthetic_source{0,1} ,
enantiomer ,
sequence ,
ec_number*
Children details ec_number enantiomer molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression sequence synthetic_source
Instance
<protein_or_peptide chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<recombinant_expression database= "" > {0,1} </recombinant_expression>
<synthetic_source database= "" > {0,1} </synthetic_source>
<enantiomer > {1,1} </enantiomer>
<sequence > {1,1} </sequence>
<ec_number > {0,unbounded} </ec_number>
</protein_or_peptide>
Attributes
Source
<xs:element name= "protein_or_peptide" type= "protein_or_peptide_macromolecule_type" substitutionGroup= "macromolecule" />
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
Element rna
Namespace No namespace
Diagram
Type rna_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence ,
classification{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1} ,
ec_number*
Children classification details ec_number molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag sequence structure synthetic_flag synthetic_source
Instance
<rna chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<sequence > {1,1} </sequence>
<classification > {0,1} </classification>
<structure > {0,1} </structure>
<synthetic_flag > {0,1} </synthetic_flag>
<synthetic_source database= "" > {0,1} </synthetic_source>
<ec_number > {0,unbounded} </ec_number>
</rna>
Attributes
Source
<xs:element name= "rna" type= "rna_macromolecule_type" substitutionGroup= "macromolecule" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
MESSENGER
enumeration
TRANSFER
enumeration
RIBOSOMAL
enumeration
NON-CODING
enumeration
INTERFERENCE
enumeration
SMALL INTERFERENCE
enumeration
GENOMIC
enumeration
PRE-MESSENGER
enumeration
SMALL NUCLEOLAR
enumeration
TRANSFER-MESSENGER
enumeration
OTHER
Source
<xs:element name= "classification" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "MESSENGER" />
<xs:enumeration value= "TRANSFER" />
<xs:enumeration value= "RIBOSOMAL" />
<xs:enumeration value= "NON-CODING" />
<xs:enumeration value= "INTERFERENCE" />
<xs:enumeration value= "SMALL INTERFERENCE" />
<xs:enumeration value= "GENOMIC" />
<xs:enumeration value= "PRE-MESSENGER" />
<xs:enumeration value= "SMALL NUCLEOLAR" />
<xs:enumeration value= "TRANSFER-MESSENGER" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type macromolecule_source_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Instance
<synthetic_source database= "" >
<organism ncbi= "" > {1,1} </organism>
<strain > {0,1} </strain>
<synonym_organism > {0,1} </synonym_organism>
<organ > {0,1} </organ>
<tissue > {0,1} </tissue>
<cell > {0,1} </cell>
<organelle > {0,1} </organelle>
<cellular_location > {0,1} </cellular_location>
</synthetic_source>
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
Element saccharide
Namespace No namespace
Diagram
Type saccharide_macromolecule_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
enantiomer ,
formula{0,1} ,
external_references*
Children details enantiomer external_references formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag
Instance
<saccharide chimera= "" macromolecule_id= "" mutant= "" >
<name synonym= "" > {1,1} </name>
<natural_source database= "" > {0,1} </natural_source>
<molecular_weight > {0,1} </molecular_weight>
<details > {0,1} </details>
<number_of_copies > {0,1} </number_of_copies>
<oligomeric_state > {0,1} </oligomeric_state>
<recombinant_exp_flag > {0,1} </recombinant_exp_flag>
<enantiomer > {1,1} </enantiomer>
<formula > {0,1} </formula>
<external_references type= "" > {0,unbounded} </external_references>
</saccharide>
Attributes
Source
<xs:element name= "saccharide" type= "saccharide_macromolecule_type" substitutionGroup= "macromolecule" />
Element crystallography_preparation
Namespace No namespace
Diagram
Type crystallography_preparation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1} ,
crystal_formation{0,1}
Children buffer concentration crystal_formation details grid shadowing staining sugar_embedding vitrification
Instance
<crystallography_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
<crystal_formation > {0,1} </crystal_formation>
</crystallography_preparation>
Attributes
Source
<xs:element name= "crystallography_preparation" type= "crystallography_preparation_type" substitutionGroup= "specimen_preparation" />
Element helical_preparation
Namespace No namespace
Diagram
Type helical_preparation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Instance
<helical_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
</helical_preparation>
Attributes
Source
<xs:element name= "helical_preparation" type= "helical_preparation_type" substitutionGroup= "specimen_preparation" />
Element single_particle_preparation
Namespace No namespace
Diagram
Type single_particle_preparation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Instance
<single_particle_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
</single_particle_preparation>
Attributes
Source
<xs:element name= "single_particle_preparation" type= "single_particle_preparation_type" substitutionGroup= "specimen_preparation" />
Element subtomogram_averaging_preparation
Namespace No namespace
Diagram
Type subtomogram_averaging_preparation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Instance
<subtomogram_averaging_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
</subtomogram_averaging_preparation>
Attributes
Source
<xs:element name= "subtomogram_averaging_preparation" type= "subtomogram_averaging_preparation_type" substitutionGroup= "specimen_preparation" />
Element tomography_preparation
Namespace No namespace
Diagram
Type tomography_preparation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1} ,
fiducial_markers_list{0,1} ,
high_pressure_freezing{0,1} ,
embedding_material{0,1} ,
cryo_protectant{0,1} ,
sectioning{0,1}
Children buffer concentration cryo_protectant details embedding_material fiducial_markers_list grid high_pressure_freezing sectioning shadowing staining sugar_embedding vitrification
Instance
<tomography_preparation preparation_id= "" >
<concentration units= "" > {0,1} </concentration>
<buffer > {0,1} </buffer>
<staining > {0,1} </staining>
<sugar_embedding > {0,1} </sugar_embedding>
<shadowing > {0,1} </shadowing>
<grid > {0,1} </grid>
<vitrification > {0,1} </vitrification>
<details > {0,1} </details>
<fiducial_markers_list > {0,1} </fiducial_markers_list>
<high_pressure_freezing > {0,1} </high_pressure_freezing>
<embedding_material > {0,1} </embedding_material>
<cryo_protectant > {0,1} </cryo_protectant>
<sectioning > {0,1} </sectioning>
</tomography_preparation>
Attributes
Source
<xs:element name= "tomography_preparation" type= "tomography_preparation_type" substitutionGroup= "specimen_preparation" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
BAL-TEC HPM 010
enumeration
EMS-002 RAPID IMMERSION FREEZER
enumeration
LEICA EM HPM100
enumeration
LEICA EM PACT
enumeration
LEICA EM PACT2
enumeration
OTHER
Source
<xs:element name= "instrument" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "BAL-TEC HPM 010" />
<xs:enumeration value= "EMS-002 RAPID IMMERSION FREEZER" />
<xs:enumeration value= "LEICA EM HPM100" />
<xs:enumeration value= "LEICA EM PACT" />
<xs:enumeration value= "LEICA EM PACT2" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Properties
Model
instrument ,
ion ,
voltage ,
current ,
dose_rate{0,1} ,
duration ,
temperature ,
initial_thickness ,
final_thickness ,
details{0,1}
Children current details dose_rate duration final_thickness initial_thickness instrument ion temperature voltage
Instance
<focused_ion_beam >
<instrument > {1,1} </instrument>
<ion > {1,1} </ion>
<voltage units= "kV" > {1,1} </voltage>
<current units= "nA" > {1,1} </current>
<dose_rate units= "ions/(cm^2*s)" > {0,1} </dose_rate>
<duration units= "s" > {1,1} </duration>
<temperature units= "K" > {1,1} </temperature>
<initial_thickness units= "nm" > {1,1} </initial_thickness>
<final_thickness units= "nm" > {1,1} </final_thickness>
<details > {0,1} </details>
</focused_ion_beam>
Source
<xs:element name= "focused_ion_beam" >
<xs:complexType >
<xs:sequence >
<xs:element name= "instrument" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "DB235" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "ion" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "GALLIUM+" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "voltage" type= "fib_voltage_type" />
<xs:element name= "current" type= "fib_current_type" />
<xs:element name= "dose_rate" type= "fib_dose_rate_type" minOccurs= "0" />
<xs:element name= "duration" type= "fib_duration_type" />
<xs:element name= "temperature" type= "temperature_type" />
<xs:element name= "initial_thickness" type= "fib_initial_thickness_type" />
<xs:element name= "final_thickness" type= "fib_final_thickness_type" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
Element crystallography_microscopy
Namespace No namespace
Diagram
Type crystallography_microscopy_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1} ,
camera_length , (
tilt_list{0,1} |
tilt_series* )
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification camera_length cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min tilt_list tilt_series
Instance
<crystallography_microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
<camera_length units= "mm" > {1,1} </camera_length>
<tilt_list > {0,1} </tilt_list>
<tilt_series > {0,unbounded} </tilt_series>
</crystallography_microscopy>
Attributes
Source
<xs:element name= "crystallography_microscopy" type= "crystallography_microscopy_type" substitutionGroup= "microscopy" />
Element helical_microscopy
Namespace No namespace
Diagram
Type helical_microscopy_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Instance
<helical_microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
</helical_microscopy>
Attributes
Source
<xs:element name= "helical_microscopy" type= "helical_microscopy_type" substitutionGroup= "microscopy" />
Element single_particle_microscopy
Namespace No namespace
Diagram
Type single_particle_microscopy_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Instance
<single_particle_microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
</single_particle_microscopy>
Attributes
Source
<xs:element name= "single_particle_microscopy" type= "single_particle_microscopy_type" substitutionGroup= "microscopy" />
Element subtomogram_averaging_microscopy
Namespace No namespace
Diagram
Type tomography_microscopy_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1} ,
tilt_series*
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min tilt_series
Instance
<subtomogram_averaging_microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
<tilt_series > {0,unbounded} </tilt_series>
</subtomogram_averaging_microscopy>
Attributes
Source
<xs:element name= "subtomogram_averaging_microscopy" type= "tomography_microscopy_type" substitutionGroup= "microscopy" />
Element tomography_microscopy
Namespace No namespace
Diagram
Type tomography_microscopy_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1} ,
tilt_series*
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min tilt_series
Instance
<tomography_microscopy microscopy_id= "" >
<specimen_preparations > {0,1} </specimen_preparations>
<microscope > {1,1} </microscope>
<illumination_mode > {1,1} </illumination_mode>
<imaging_mode > {1,1} </imaging_mode>
<electron_source > {1,1} </electron_source>
<acceleration_voltage units= "kV" > {1,1} </acceleration_voltage>
<c2_aperture_diameter units= "µm" > {0,1} </c2_aperture_diameter>
<nominal_cs units= "mm" > {0,1} </nominal_cs>
<nominal_defocus_min units= "µm" > {0,1} </nominal_defocus_min>
<calibrated_defocus_min units= "µm" > {0,1} </calibrated_defocus_min>
<nominal_defocus_max units= "µm" > {0,1} </nominal_defocus_max>
<calibrated_defocus_max units= "µm" > {0,1} </calibrated_defocus_max>
<nominal_magnification > {0,1} </nominal_magnification>
<calibrated_magnification > {0,1} </calibrated_magnification>
<specimen_holder_model > {0,1} </specimen_holder_model>
<cooling_holder_cryogen > {0,1} </cooling_holder_cryogen>
<temperature > {0,1} </temperature>
<alignment_procedure > {0,1} </alignment_procedure>
<specialist_optics > {0,1} </specialist_optics>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<date > {0,1} </date>
<image_recording_list > {1,1} </image_recording_list>
<specimen_holder > {0,1} </specimen_holder>
<tilt_angle_min > {0,1} </tilt_angle_min>
<tilt_angle_max > {0,1} </tilt_angle_max>
<tilt_series > {0,unbounded} </tilt_series>
</tomography_microscopy>
Attributes
Source
<xs:element name= "tomography_microscopy" type= "tomography_microscopy_type" substitutionGroup= "microscopy" />
Element crystallography_processing
Namespace No namespace
Diagram
Type crystallography_processing_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
crystal_parameters{0,1} ,
startup_model* ,
ctf_correction{0,1} ,
molecular_replacement{0,1} ,
lattice_distortion_correction_software_list{0,1} ,
symmetry_determination_software_list{0,1} ,
merging_software_list{0,1} ,
crystallography_statistics{0,1}
Children crystal_parameters crystallography_statistics ctf_correction details final_reconstruction image_recording_id lattice_distortion_correction_software_list merging_software_list molecular_replacement startup_model symmetry_determination_software_list
Instance
<crystallography_processing image_processing_id= "" >
<image_recording_id > {1,1} </image_recording_id>
<details > {0,1} </details>
<final_reconstruction > {0,1} </final_reconstruction>
<crystal_parameters > {0,1} </crystal_parameters>
<startup_model type_of_model= "" > {0,unbounded} </startup_model>
<ctf_correction > {0,1} </ctf_correction>
<molecular_replacement > {0,1} </molecular_replacement>
<lattice_distortion_correction_software_list > {0,1} </lattice_distortion_correction_software_list>
<symmetry_determination_software_list > {0,1} </symmetry_determination_software_list>
<merging_software_list > {0,1} </merging_software_list>
<crystallography_statistics > {0,1} </crystallography_statistics>
</crystallography_processing>
Attributes
Source
<xs:element name= "crystallography_processing" substitutionGroup= "image_processing" type= "crystallography_processing_type" />
Namespace No namespace
Diagram
Type non_subtom_final_reconstruction_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_images_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_images_used reconstruction_filtering resolution resolution_method software_list
Instance
<final_reconstruction >
<number_classes_used > {0,1} </number_classes_used>
<applied_symmetry > {0,1} </applied_symmetry>
<algorithm > {0,1} </algorithm>
<resolution res_type= "" units= "Å" > {0,1} </resolution>
<resolution_method > {0,1} </resolution_method>
<reconstruction_filtering > {0,1} </reconstruction_filtering>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<number_images_used > {0,1} </number_images_used>
</final_reconstruction>
Source
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type reconstruction_algorithm_type
Properties
content
simple
minOccurs
0
Facets
enumeration
ALGEBRAIC (ARTS)
enumeration
BACK PROJECTION
enumeration
EXACT BACK PROJECTION
enumeration
FOURIER SPACE
enumeration
SIMULTANEOUS ITERATIVE (SIRT)
Source
<xs:element name= "algorithm" type= "reconstruction_algorithm_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
content
simple
minOccurs
0
Facets
enumeration
DIFFRACTION PATTERN/LAYERLINES
enumeration
FSC 0.143 CUT-OFF
enumeration
FSC 0.33 CUT-OFF
enumeration
FSC 0.5 CUT-OFF
enumeration
FSC 1/2 BIT CUT-OFF
enumeration
FSC 3 SIGMA CUT-OFF
enumeration
OTHER
Source
<xs:element name= "resolution_method" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "DIFFRACTION PATTERN/LAYERLINES" />
<xs:enumeration value= "FSC 0.143 CUT-OFF" />
<xs:enumeration value= "FSC 0.33 CUT-OFF" />
<xs:enumeration value= "FSC 0.5 CUT-OFF" />
<xs:enumeration value= "FSC 1/2 BIT CUT-OFF" />
<xs:enumeration value= "FSC 3 SIGMA CUT-OFF" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
SPHERE
enumeration
SOFT SPHERE
enumeration
GAUSSIAN
enumeration
CIRCLE
enumeration
RECTANGLE
enumeration
CYLINDER
enumeration
OTHER
Source
<xs:element name= "geometrical_shape" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "SPHERE" />
<xs:enumeration value= "SOFT SPHERE" />
<xs:enumeration value= "GAUSSIAN" />
<xs:enumeration value= "CIRCLE" />
<xs:enumeration value= "RECTANGLE" />
<xs:enumeration value= "CYLINDER" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type unit_cell_type
Properties
Model
Children a alpha b beta c c_sampling_length gamma
Instance
<unit_cell >
<a units= "Å" > {1,1} </a>
<b units= "Å" > {1,1} </b>
<c units= "Å" > {1,1} </c>
<c_sampling_length units= "Å" > {0,1} </c_sampling_length>
<gamma units= "deg" > {1,1} </gamma>
<alpha units= "deg" > {0,1} </alpha>
<beta units= "deg" > {0,1} </beta>
</unit_cell>
Source
<xs:element name= "unit_cell" type= "unit_cell_type" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
C 1 2
enumeration
C 2 2 2
enumeration
P 1
enumeration
P 1 2
enumeration
P 1 21
enumeration
P 2
enumeration
P 2 2 2
enumeration
P 2 2 21
enumeration
P 2 21 21
enumeration
P 3
enumeration
P 3 1 2
enumeration
P 3 2 1
enumeration
P 4
enumeration
P 4 2 2
enumeration
P 4 21 2
enumeration
P 6
enumeration
P 6 2 2
Source
<xs:element name= "plane_group" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "C 1 2" />
<xs:enumeration value= "C 2 2 2" />
<xs:enumeration value= "P 1" />
<xs:enumeration value= "P 1 2" />
<xs:enumeration value= "P 1 21" />
<xs:enumeration value= "P 2" />
<xs:enumeration value= "P 2 2 2" />
<xs:enumeration value= "P 2 2 21" />
<xs:enumeration value= "P 2 21 21" />
<xs:enumeration value= "P 3" />
<xs:enumeration value= "P 3 1 2" />
<xs:enumeration value= "P 3 2 1" />
<xs:enumeration value= "P 4" />
<xs:enumeration value= "P 4 2 2" />
<xs:enumeration value= "P 4 21 2" />
<xs:enumeration value= "P 6" />
<xs:enumeration value= "P 6 2 2" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Namespace No namespace
Diagram
Type starting_map_type
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
Children details emdb_id insilico_model orthogonal_tilt other pdb_model random_conical_tilt
Instance
<startup_model type_of_model= "" >
<random_conical_tilt > {0,1} </random_conical_tilt>
<orthogonal_tilt > {0,1} </orthogonal_tilt>
<emdb_id > {0,1} </emdb_id>
<pdb_model > {0,1} </pdb_model>
<insilico_model > {0,1} </insilico_model>
<other > {0,1} </other>
<details > {0,1} </details>
</startup_model>
Attributes
Source
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type crystallography_statistics_type
Properties
content
complex
minOccurs
0
Model
number_intensities_measured ,
number_structure_factors ,
fourier_space_coverage ,
r_sym{0,1} ,
r_merge ,
overall_phase_error{0,1} ,
overall_phase_residual{0,1} ,
phase_error_rejection_criteria ,
high_resolution ,
shell_list{0,1} ,
details{0,1}
Children details fourier_space_coverage high_resolution number_intensities_measured number_structure_factors overall_phase_error overall_phase_residual phase_error_rejection_criteria r_merge r_sym shell_list
Instance
<crystallography_statistics >
<number_intensities_measured > {1,1} </number_intensities_measured>
<number_structure_factors > {1,1} </number_structure_factors>
<fourier_space_coverage > {1,1} </fourier_space_coverage>
<r_sym > {0,1} </r_sym>
<r_merge > {1,1} </r_merge>
<overall_phase_error > {0,1} </overall_phase_error>
<overall_phase_residual > {0,1} </overall_phase_residual>
<phase_error_rejection_criteria > {1,1} </phase_error_rejection_criteria>
<high_resolution units= "Å" > {1,1} </high_resolution>
<shell_list > {0,1} </shell_list>
<details > {0,1} </details>
</crystallography_statistics>
Source
<xs:element name= "crystallography_statistics" type= "crystallography_statistics_type" minOccurs= "0" />
Namespace No namespace
Diagram
Properties
content
complex
maxOccurs
unbounded
Model
Children fourier_space_coverage high_resolution low_resolution multiplicity number_structure_factors phase_residual
Instance
<shell shell_id= "" >
<high_resolution units= "Å" > {1,1} </high_resolution>
<low_resolution units= "Å" > {1,1} </low_resolution>
<number_structure_factors > {1,1} </number_structure_factors>
<phase_residual > {1,1} </phase_residual>
<fourier_space_coverage > {1,1} </fourier_space_coverage>
<multiplicity > {1,1} </multiplicity>
</shell>
Attributes
QName
Type
Use
shell_id xs:positiveInteger optional
Source
<xs:element name= "shell" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "high_resolution" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "low_resolution" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "number_structure_factors" type= "xs:positiveInteger" />
<xs:element name= "phase_residual" type= "xs:float" />
<xs:element name= "fourier_space_coverage" type= "xs:float" />
<xs:element name= "multiplicity" type= "xs:float" />
</xs:sequence>
<xs:attribute name= "shell_id" type= "xs:positiveInteger" />
</xs:complexType>
</xs:element>
Element helical_processing
Namespace No namespace
Diagram
Type helical_processing_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
ctf_correction{0,1} ,
segment_selection* ,
refinement{0,1} ,
startup_model* ,
helical_layer_lines{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details final_angle_assignment final_reconstruction helical_layer_lines image_recording_id initial_angle_assignment refinement segment_selection startup_model
Instance
<helical_processing image_processing_id= "" >
<image_recording_id > {1,1} </image_recording_id>
<details > {0,1} </details>
<final_reconstruction > {0,1} </final_reconstruction>
<ctf_correction > {0,1} </ctf_correction>
<segment_selection > {0,unbounded} </segment_selection>
<refinement > {0,1} </refinement>
<startup_model type_of_model= "" > {0,unbounded} </startup_model>
<helical_layer_lines > {0,1} </helical_layer_lines>
<initial_angle_assignment > {0,1} </initial_angle_assignment>
<final_angle_assignment > {0,1} </final_angle_assignment>
<crystal_parameters > {0,1} </crystal_parameters>
</helical_processing>
Attributes
Source
<xs:element name= "helical_processing" substitutionGroup= "image_processing" type= "helical_processing_type" />
Namespace No namespace
Diagram
Type non_subtom_final_reconstruction_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_images_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_images_used reconstruction_filtering resolution resolution_method software_list
Instance
<final_reconstruction >
<number_classes_used > {0,1} </number_classes_used>
<applied_symmetry > {0,1} </applied_symmetry>
<algorithm > {0,1} </algorithm>
<resolution res_type= "" units= "Å" > {0,1} </resolution>
<resolution_method > {0,1} </resolution_method>
<reconstruction_filtering > {0,1} </reconstruction_filtering>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<number_images_used > {0,1} </number_images_used>
</final_reconstruction>
Source
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type segment_selection_type
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
number_selected{0,1} ,
segment_length{0,1} ,
segment_overlap{0,1} ,
total_filament_length{0,1} ,
software_list{0,1} ,
details{0,1}
Children details number_selected segment_length segment_overlap software_list total_filament_length
Instance
<segment_selection >
<number_selected > {0,1} </number_selected>
<segment_length units= "Å" > {0,1} </segment_length>
<segment_overlap units= "Å" > {0,1} </segment_overlap>
<total_filament_length units= "Å" > {0,1} </total_filament_length>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
</segment_selection>
Source
<xs:element name= "segment_selection" type= "segment_selection_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type starting_map_type
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
Children details emdb_id insilico_model orthogonal_tilt other pdb_model random_conical_tilt
Instance
<startup_model type_of_model= "" >
<random_conical_tilt > {0,1} </random_conical_tilt>
<orthogonal_tilt > {0,1} </orthogonal_tilt>
<emdb_id > {0,1} </emdb_id>
<pdb_model > {0,1} </pdb_model>
<insilico_model > {0,1} </insilico_model>
<other > {0,1} </other>
<details > {0,1} </details>
</startup_model>
Attributes
Source
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type starting_map_type
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
Children details emdb_id insilico_model orthogonal_tilt other pdb_model random_conical_tilt
Instance
<startup_model type_of_model= "" >
<random_conical_tilt > {0,1} </random_conical_tilt>
<orthogonal_tilt > {0,1} </orthogonal_tilt>
<emdb_id > {0,1} </emdb_id>
<pdb_model > {0,1} </pdb_model>
<insilico_model > {0,1} </insilico_model>
<other > {0,1} </other>
<details > {0,1} </details>
</startup_model>
Attributes
Source
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type restriction of xs:token
Properties
Facets
enumeration
ANGULAR RECONSTITUTION
enumeration
COMMON LINE
enumeration
NOT APPLICABLE
enumeration
OTHER
enumeration
PROJECTION MATCHING
enumeration
RANDOM ASSIGNMENT
enumeration
MAXIMUM LIKELIHOOD
Source
<xs:element name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "ANGULAR RECONSTITUTION" />
<xs:enumeration value= "COMMON LINE" />
<xs:enumeration value= "NOT APPLICABLE" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PROJECTION MATCHING" />
<xs:enumeration value= "RANDOM ASSIGNMENT" />
<xs:enumeration value= "MAXIMUM LIKELIHOOD" />
</xs:restriction>
</xs:simpleType>
</xs:element>
Element singleparticle_processing
Namespace No namespace
Diagram
Type singleparticle_processing_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
image_recording_id ,
details{0,1} ,
particle_selection* ,
ctf_correction{0,1} ,
startup_model* ,
final_reconstruction{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
final_multi_reference_alignment{0,1} ,
final_two_d_classification{0,1} ,
final_three_d_classification{0,1}
Children ctf_correction details final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification final_two_d_classification image_recording_id initial_angle_assignment particle_selection startup_model
Instance
<singleparticle_processing image_processing_id= "" >
<image_recording_id > {1,1} </image_recording_id>
<details > {0,1} </details>
<particle_selection > {0,unbounded} </particle_selection>
<ctf_correction > {0,1} </ctf_correction>
<startup_model type_of_model= "" > {0,unbounded} </startup_model>
<final_reconstruction > {0,1} </final_reconstruction>
<initial_angle_assignment > {0,1} </initial_angle_assignment>
<final_angle_assignment > {0,1} </final_angle_assignment>
<final_multi_reference_alignment > {0,1} </final_multi_reference_alignment>
<final_two_d_classification > {0,1} </final_two_d_classification>
<final_three_d_classification > {0,1} </final_three_d_classification>
</singleparticle_processing>
Attributes
Source
<xs:element name= "singleparticle_processing" substitutionGroup= "image_processing" type= "singleparticle_processing_type" />
Namespace No namespace
Diagram
Type starting_map_type
Properties
content
complex
minOccurs
0
maxOccurs
unbounded
Model
Children details emdb_id insilico_model orthogonal_tilt other pdb_model random_conical_tilt
Instance
<startup_model type_of_model= "" >
<random_conical_tilt > {0,1} </random_conical_tilt>
<orthogonal_tilt > {0,1} </orthogonal_tilt>
<emdb_id > {0,1} </emdb_id>
<pdb_model > {0,1} </pdb_model>
<insilico_model > {0,1} </insilico_model>
<other > {0,1} </other>
<details > {0,1} </details>
</startup_model>
Attributes
Source
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
Namespace No namespace
Diagram
Type non_subtom_final_reconstruction_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_images_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_images_used reconstruction_filtering resolution resolution_method software_list
Instance
<final_reconstruction >
<number_classes_used > {0,1} </number_classes_used>
<applied_symmetry > {0,1} </applied_symmetry>
<algorithm > {0,1} </algorithm>
<resolution res_type= "" units= "Å" > {0,1} </resolution>
<resolution_method > {0,1} </resolution_method>
<reconstruction_filtering > {0,1} </reconstruction_filtering>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<number_images_used > {0,1} </number_images_used>
</final_reconstruction>
Source
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
Element subtomogram_averaging_processing
Namespace No namespace
Diagram
Type subtomogram_averaging_processing_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
extraction ,
ctf_correction{0,1} ,
final_multi_reference_alignment{0,1} ,
final_three_d_classification{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details extraction final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification image_recording_id
Instance
<subtomogram_averaging_processing image_processing_id= "" >
<image_recording_id > {1,1} </image_recording_id>
<details > {0,1} </details>
<final_reconstruction > {0,1} </final_reconstruction>
<extraction > {1,1} </extraction>
<ctf_correction > {0,1} </ctf_correction>
<final_multi_reference_alignment > {0,1} </final_multi_reference_alignment>
<final_three_d_classification > {0,1} </final_three_d_classification>
<final_angle_assignment > {0,1} </final_angle_assignment>
<crystal_parameters > {0,1} </crystal_parameters>
</subtomogram_averaging_processing>
Attributes
Source
<xs:element name= "subtomogram_averaging_processing" substitutionGroup= "image_processing" type= "subtomogram_averaging_processing_type" />
Namespace No namespace
Diagram
Type subtomogram_final_reconstruction_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_subtomograms_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_subtomograms_used reconstruction_filtering resolution resolution_method software_list
Instance
<final_reconstruction >
<number_classes_used > {0,1} </number_classes_used>
<applied_symmetry > {0,1} </applied_symmetry>
<algorithm > {0,1} </algorithm>
<resolution res_type= "" units= "Å" > {0,1} </resolution>
<resolution_method > {0,1} </resolution_method>
<reconstruction_filtering > {0,1} </reconstruction_filtering>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<number_subtomograms_used > {0,1} </number_subtomograms_used>
</final_reconstruction>
Source
<xs:element name= "final_reconstruction" type= "subtomogram_final_reconstruction_type" minOccurs= "0" />
Namespace No namespace
Diagram
Type xs:positiveInteger
Properties
Source
Namespace No namespace
Diagram
Type xs:positiveInteger
Properties
Source
Namespace No namespace
Diagram
Type xs:token
Properties
Source
Namespace No namespace
Diagram
Type xs:string
Properties
Source
Namespace No namespace
Diagram
Type xs:string
Properties
Source
Element tomography_processing
Namespace No namespace
Diagram
Type tomography_processing_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
series_aligment_software_list{0,1} ,
ctf_correction{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details final_reconstruction image_recording_id series_aligment_software_list
Instance
<tomography_processing image_processing_id= "" >
<image_recording_id > {1,1} </image_recording_id>
<details > {0,1} </details>
<final_reconstruction > {0,1} </final_reconstruction>
<series_aligment_software_list > {0,1} </series_aligment_software_list>
<ctf_correction > {0,1} </ctf_correction>
<crystal_parameters > {0,1} </crystal_parameters>
</tomography_processing>
Attributes
Source
<xs:element name= "tomography_processing" substitutionGroup= "image_processing" type= "tomography_processing_type" />
Namespace No namespace
Diagram
Type non_subtom_final_reconstruction_type
Type hierarchy
Properties
content
complex
minOccurs
0
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_images_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_images_used reconstruction_filtering resolution resolution_method software_list
Instance
<final_reconstruction >
<number_classes_used > {0,1} </number_classes_used>
<applied_symmetry > {0,1} </applied_symmetry>
<algorithm > {0,1} </algorithm>
<resolution res_type= "" units= "Å" > {0,1} </resolution>
<resolution_method > {0,1} </resolution_method>
<reconstruction_filtering > {0,1} </reconstruction_filtering>
<software_list > {0,1} </software_list>
<details > {0,1} </details>
<number_images_used > {0,1} </number_images_used>
</final_reconstruction>
Source
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
Element crystallography_validation
Namespace No namespace
Diagram
Type crystallography_validation_type
Type hierarchy
Properties
Substitution Group Affiliation
Model
Children data_completeness details file number_observed_reflections number_unique_reflections parallel_resolution perpendicular_resolution weighted_phase_residual weighted_r_factor
Instance
<crystallography_validation >
<file > {1,1} </file>
<details > {0,1} </details>
<parallel_resolution > {1,1} </parallel_resolution>
<perpendicular_resolution > {1,1} </perpendicular_resolution>
<number_observed_reflections > {1,1} </number_observed_reflections>
<number_unique_reflections > {1,1} </number_unique_reflections>
<weighted_phase_residual > {1,1} </weighted_phase_residual>
<weighted_r_factor > {1,1} </weighted_r_factor>
<data_completeness > {1,1} </data_completeness>
</crystallography_validation>
Source
<xs:element name= "crystallography_validation" substitutionGroup= "validation_method" type= "crystallography_validation_type" />
Element fsc_curve
Element layer_lines
Element structure_factors
Complex Type entry_type
Complex Type admin_type
Namespace No namespace
Diagram
Used by
Model
status_history_list{0,1} ,
current_status ,
sites ,
key_dates ,
obsolete_list{0,1} ,
superseded_by_list{0,1} ,
grant_support{0,1} ,
microscopy_center{0,1} ,
contact_author* ,
title ,
authors_list ,
details{0,1} ,
keywords{0,1} ,
replace_existing_entry{0,1}
Children authors_list contact_author current_status details grant_support key_dates keywords microscopy_center obsolete_list replace_existing_entry sites status_history_list superseded_by_list title
Source
<xs:complexType name= "admin_type" >
<xs:sequence >
<xs:element name= "status_history_list" type= "version_list_type" minOccurs= "0" />
<xs:element name= "current_status" type= "version_type" />
<xs:element name= "sites" >
<xs:complexType >
<xs:sequence >
<xs:element name= "deposition" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PDBe" />
<xs:enumeration value= "PDBj" />
<xs:enumeration value= "RCSB" />
<xs:enumeration value= "PDBc" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "last_processing" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PDBe" />
<xs:enumeration value= "PDBj" />
<xs:enumeration value= "RCSB" />
<xs:enumeration value= "PDBc" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "key_dates" >
<xs:complexType >
<xs:sequence >
<xs:element name= "deposition" type= "xs:date" />
<xs:element name= "header_release" type= "xs:date" minOccurs= "0" />
<xs:element name= "map_release" type= "xs:date" minOccurs= "0" />
<xs:element name= "obsolete" type= "xs:date" minOccurs= "0" />
<xs:element name= "update" type= "xs:date" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "obsolete_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "entry" type= "supersedes_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "superseded_by_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "entry" type= "supersedes_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "grant_support" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "grant_reference" type= "grant_reference_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "microscopy_center" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "name" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "MICRO_CENTER_1" />
<xs:enumeration value= "MICRO_CENTER_2" />
<xs:enumeration value= "MICRO_CENTER_3" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "country" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "UK" />
<xs:enumeration value= "USA" />
<xs:enumeration value= "Japan" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "contact_author" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:complexContent >
<xs:extension base= "contact_details_type" >
<xs:attribute name= "private" fixed= "true" use= "required" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
</xs:element>
<xs:element name= "title" type= "xs:token" />
<xs:element name= "authors_list" >
<xs:complexType >
<xs:sequence >
<xs:element name= "author" type= "author_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:token" minOccurs= "0" />
<xs:element name= "keywords" type= "xs:string" minOccurs= "0" />
<xs:element name= "replace_existing_entry" type= "xs:boolean" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Complex Type version_list_type
Complex Type version_type
Complex Type code_type
Simple Type status_code_type
Namespace No namespace
Diagram
Type restriction of xs:token
Facets
enumeration
AUCO
enumeration
AUTH
enumeration
AUXS
enumeration
AUXU
enumeration
HOLD
enumeration
HOLD8W
enumeration
HPUB
enumeration
OBS
enumeration
POLC
enumeration
PROC
enumeration
REFI
enumeration
REL
enumeration
REPL
enumeration
REUP
enumeration
WAIT
enumeration
WDRN
Used by
Source
<xs:simpleType name= "status_code_type" >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "AUCO" />
<xs:enumeration value= "AUTH" />
<xs:enumeration value= "AUXS" />
<xs:enumeration value= "AUXU" />
<xs:enumeration value= "HOLD" />
<xs:enumeration value= "HOLD8W" />
<xs:enumeration value= "HPUB" />
<xs:enumeration value= "OBS" />
<xs:enumeration value= "POLC" />
<xs:enumeration value= "PROC" />
<xs:enumeration value= "REFI" />
<xs:enumeration value= "REL" />
<xs:enumeration value= "REPL" />
<xs:enumeration value= "REUP" />
<xs:enumeration value= "WAIT" />
<xs:enumeration value= "WDRN" />
</xs:restriction>
</xs:simpleType>
Complex Type supersedes_type
Simple Type emdb_id_type
Complex Type grant_reference_type
Complex Type contact_details_type
Namespace No namespace
Diagram
Used by
Model
Children country email fax first_name last_name middle_name organization post_or_zip_code role state_or_province street telephone title town_or_city
Source
Complex Type telephone_number_type
Simple Type author_type
Complex Type crossreferences_type
Complex Type emdb_cross_reference_list_type
Complex Type emdb_cross_reference_type
Complex Type pdb_cross_reference_list_type
Complex Type pdb_cross_reference_type
Simple Type pdb_code_type
Complex Type auxiliary_link_type
Complex Type sample_type
Complex Type sci_name_type
Complex Type base_supramolecule_type
Namespace No namespace
Diagram
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1}
Children category details external_references macromolecule_list name number_of_copies oligomeric_state parent recombinant_exp_flag
Attributes
Source
<xs:complexType name= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "name" type= "sci_name_type" />
<xs:element name= "category" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "complex_category_type" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "GO" />
<xs:enumeration value= "ARBITRARY DEFINITION" />
<xs:enumeration value= "PROTEIN ONTOLOGY" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "parent" type= "xs:nonNegativeInteger" />
<xs:element name= "macromolecule_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "macromolecule" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "macromolecule_id" type= "xs:positiveInteger" />
<xs:element name= "number_of_copies" type= "xs:positiveInteger" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "number_of_copies" type= "pos_int_or_string_type" minOccurs= "0" />
<xs:element name= "oligomeric_state" type= "pos_int_or_string_type" minOccurs= "0" />
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "UNIPROTKB" />
<xs:enumeration value= "UNIPARC" />
<xs:enumeration value= "INTERPRO" />
<xs:enumeration value= "GO" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "recombinant_exp_flag" type= "xs:boolean" minOccurs= "0" maxOccurs= "1" />
</xs:sequence>
<xs:attribute name= "supramolecule_id" type= "xs:positiveInteger" use= "required" />
</xs:complexType>
Simple Type complex_category_type
Simple Type pos_int_or_string_type
Complex Type macromolecule_list_type
Complex Type base_macromolecule_type
Namespace No namespace
Diagram
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1}
Children details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag
Attributes
Source
<xs:complexType name= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "name" type= "sci_name_type" />
<xs:element name= "natural_source" type= "macromolecule_source_type" minOccurs= "0" />
<xs:element name= "molecular_weight" type= "molecular_weight_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "number_of_copies" type= "pos_int_or_string_type" minOccurs= "0" />
<xs:element name= "oligomeric_state" type= "pos_int_or_string_type" minOccurs= "0" />
<xs:element name= "recombinant_exp_flag" type= "xs:boolean" minOccurs= "0" maxOccurs= "1" />
</xs:sequence>
<xs:attribute name= "macromolecule_id" type= "xs:positiveInteger" use= "required" />
<xs:attribute name= "mutant" type= "xs:boolean" />
<xs:attribute name= "chimera" type= "xs:boolean" />
</xs:complexType>
Complex Type macromolecule_source_type
Namespace No namespace
Diagram
Type extension of base_source_type
Type hierarchy
Used by
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:complexType name= "macromolecule_source_type" >
<xs:complexContent >
<xs:extension base= "base_source_type" >
<xs:sequence >
<xs:element name= "organ" type= "xs:token" minOccurs= "0" />
<xs:element name= "tissue" type= "xs:token" minOccurs= "0" />
<xs:element name= "cell" type= "xs:token" minOccurs= "0" />
<xs:element name= "organelle" type= "xs:token" minOccurs= "0" />
<xs:element name= "cellular_location" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type base_source_type
Complex Type organism_type
Complex Type molecular_weight_type
Simple Type allowed_assembly_weights
Complex Type structure_determination_type
Complex Type macromolecules_and_complexes_type
Complex Type base_preparation_type
Namespace No namespace
Diagram
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "base_preparation_type" >
<xs:sequence >
<xs:element name= "concentration" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_concentration" >
<xs:attribute name= "units" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "mg/mL" />
<xs:enumeration value= "mM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "buffer" type= "buffer_type" minOccurs= "0" />
<xs:element name= "staining" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "NEGATIVE" />
<xs:enumeration value= "NONE" />
<xs:enumeration value= "POSITIVE" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "material" type= "xs:token" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "sugar_embedding" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "material" type= "xs:token" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "shadowing" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "material" type= "xs:token" />
<xs:element name= "angle" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_angle_shadowing" >
<xs:attribute name= "units" type= "xs:token" fixed= "deg" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "thickness" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_thickness_shadowing" >
<xs:attribute name= "units" type= "xs:token" fixed= "nm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "grid" type= "grid_type" minOccurs= "0" />
<xs:element name= "vitrification" type= "vitrification_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "preparation_id" type= "xs:positiveInteger" use= "required" />
</xs:complexType>
Simple Type allowed_concentration
Complex Type buffer_type
Complex Type buffer_component_type
Simple Type formula_type
Simple Type allowed_angle_shadowing
Simple Type allowed_thickness_shadowing
Complex Type grid_type
Namespace No namespace
Diagram
Used by
Model
model{0,1} ,
material{0,1} ,
mesh{0,1} ,
support_film* ,
pretreatment{0,1} ,
details{0,1}
Children details material mesh model pretreatment support_film
Source
<xs:complexType name= "grid_type" >
<xs:sequence >
<xs:element name= "model" type= "xs:token" minOccurs= "0" />
<xs:element name= "material" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "COPPER" />
<xs:enumeration value= "COPPER/PALLADIUM" />
<xs:enumeration value= "COPPER/RHODIUM" />
<xs:enumeration value= "GOLD" />
<xs:enumeration value= "GRAPHENE OXIDE" />
<xs:enumeration value= "MOLYBDENUM" />
<xs:enumeration value= "NICKEL" />
<xs:enumeration value= "NICKEL/TITANIUM" />
<xs:enumeration value= "PLATINUM" />
<xs:enumeration value= "SILICON NITRIDE" />
<xs:enumeration value= "TITANIUM" />
<xs:enumeration value= "TUNGSTEN" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "mesh" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "support_film" type= "film_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "pretreatment" type= "grid_pretreatment_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Complex Type film_type
Simple Type allowed_film_thickness
Complex Type grid_pretreatment_type
Simple Type allowed_time_glow_discharge
Simple Type allowed_pressure_glow_discharge
Complex Type vitrification_type
Namespace No namespace
Diagram
Used by
Model
cryogen_name ,
chamber_humidity{0,1} ,
chamber_temperature{0,1} ,
instrument{0,1} ,
details{0,1} ,
timed_resolved_state{0,1} ,
method{0,1}
Children chamber_humidity chamber_temperature cryogen_name details instrument method timed_resolved_state
Source
<xs:complexType name= "vitrification_type" >
<xs:sequence >
<xs:element name= "cryogen_name" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "ETHANE" />
<xs:enumeration value= "ETHANE-PROPANE" />
<xs:enumeration value= "FREON 12" />
<xs:enumeration value= "FREON 22" />
<xs:enumeration value= "HELIUM" />
<xs:enumeration value= "METHANE" />
<xs:enumeration value= "NITROGEN" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PROPANE" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "chamber_humidity" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_humidity_vitrification" >
<xs:attribute name= "units" type= "xs:token" fixed= "percentage" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "chamber_temperature" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_temperature_vitrification" >
<xs:attribute name= "units" type= "xs:token" fixed= "K" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "instrument" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EMS-002 RAPID IMMERSION FREEZER" />
<xs:enumeration value= "FEI VITROBOT MARK I" />
<xs:enumeration value= "FEI VITROBOT MARK II" />
<xs:enumeration value= "FEI VITROBOT MARK III" />
<xs:enumeration value= "FEI VITROBOT MARK IV" />
<xs:enumeration value= "GATAN CRYOPLUNGE 3" />
<xs:enumeration value= "HOMEMADE PLUNGER" />
<xs:enumeration value= "LEICA EM CPC" />
<xs:enumeration value= "LEICA EM GP" />
<xs:enumeration value= "LEICA KF80" />
<xs:enumeration value= "LEICA PLUNGER" />
<xs:enumeration value= "REICHERT-JUNG PLUNGER" />
<xs:enumeration value= "SPOTITON" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "timed_resolved_state" type= "xs:token" minOccurs= "0" />
<xs:element name= "method" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Simple Type allowed_humidity_vitrification
Simple Type allowed_temperature_vitrification
Complex Type base_microscopy_type
Namespace No namespace
Diagram
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Attributes
Source
<xs:complexType name= "base_microscopy_type" >
<xs:sequence >
<xs:element name= "specimen_preparations" minOccurs= "0" >
<xs:complexType >
<xs:sequence maxOccurs= "unbounded" >
<xs:element name= "specimen_preparation_id" type= "xs:positiveInteger" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "microscope" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "FEI MORGAGNI" />
<xs:enumeration value= "FEI POLARA 300" />
<xs:enumeration value= "FEI TALOS ARCTICA" />
<xs:enumeration value= "FEI TECNAI 10" />
<xs:enumeration value= "FEI TECNAI 12" />
<xs:enumeration value= "FEI TECNAI 20" />
<xs:enumeration value= "FEI TECNAI ARCTICA" />
<xs:enumeration value= "FEI TECNAI F20" />
<xs:enumeration value= "FEI TECNAI F30" />
<xs:enumeration value= "FEI TECNAI SPHERA" />
<xs:enumeration value= "FEI TECNAI SPIRIT" />
<xs:enumeration value= "FEI TITAN" />
<xs:enumeration value= "FEI TITAN KRIOS" />
<xs:enumeration value= "FEI/PHILIPS CM10" />
<xs:enumeration value= "FEI/PHILIPS CM12" />
<xs:enumeration value= "FEI/PHILIPS CM120T" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/SOPHIE" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/ST" />
<xs:enumeration value= "FEI/PHILIPS CM200FEG/UT" />
<xs:enumeration value= "FEI/PHILIPS CM200T" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/HE" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/ST" />
<xs:enumeration value= "FEI/PHILIPS CM300FEG/T" />
<xs:enumeration value= "FEI/PHILIPS EM400" />
<xs:enumeration value= "FEI/PHILIPS EM420" />
<xs:enumeration value= "HITACHI EF2000" />
<xs:enumeration value= "HITACHI H-9500SD" />
<xs:enumeration value= "HITACHI H3000 UHVEM" />
<xs:enumeration value= "HITACHI H7600" />
<xs:enumeration value= "HITACHI HF2000" />
<xs:enumeration value= "HITACHI HF3000" />
<xs:enumeration value= "JEOL 100CX" />
<xs:enumeration value= "JEOL 1000EES" />
<xs:enumeration value= "JEOL 1010" />
<xs:enumeration value= "JEOL 1200" />
<xs:enumeration value= "JEOL 1200EX" />
<xs:enumeration value= "JEOL 1200EXII" />
<xs:enumeration value= "JEOL 1230" />
<xs:enumeration value= "JEOL 1400" />
<xs:enumeration value= "JEOL 2000EX" />
<xs:enumeration value= "JEOL 2000EXII" />
<xs:enumeration value= "JEOL 2010" />
<xs:enumeration value= "JEOL 2010F" />
<xs:enumeration value= "JEOL 2010HC" />
<xs:enumeration value= "JEOL 2010HT" />
<xs:enumeration value= "JEOL 2010UHR" />
<xs:enumeration value= "JEOL 2011" />
<xs:enumeration value= "JEOL 2100" />
<xs:enumeration value= "JEOL 2100F" />
<xs:enumeration value= "JEOL 2200FS" />
<xs:enumeration value= "JEOL 2200FSC" />
<xs:enumeration value= "JEOL 3000SFF" />
<xs:enumeration value= "JEOL 3100FEF" />
<xs:enumeration value= "JEOL 3100FFC" />
<xs:enumeration value= "JEOL 3200FS" />
<xs:enumeration value= "JEOL 3200FSC" />
<xs:enumeration value= "JEOL 4000" />
<xs:enumeration value= "JEOL 4000EX" />
<xs:enumeration value= "JEOL CRYO ARM 200" />
<xs:enumeration value= "JEOL CRYO ARM 300" />
<xs:enumeration value= "JEOL KYOTO-3000SFF" />
<xs:enumeration value= "TFS GLACIOS" />
<xs:enumeration value= "TFS KRIOS" />
<xs:enumeration value= "TFS TALOS" />
<xs:enumeration value= "TFS TALOS L120C" />
<xs:enumeration value= "TFS TALOS F200C" />
<xs:enumeration value= "ZEISS LEO912" />
<xs:enumeration value= "ZEISS LIBRA120PLUS" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "illumination_mode" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "FLOOD BEAM" />
<xs:enumeration value= "SPOT SCAN" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "imaging_mode" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "BRIGHT FIELD" />
<xs:enumeration value= "DARK FIELD" />
<xs:enumeration value= "DIFFRACTION" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "electron_source" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "TUNGSTEN HAIRPIN" />
<xs:enumeration value= "LAB6" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "FIELD EMISSION GUN" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "acceleration_voltage" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_acceleration_voltage" >
<xs:attribute name= "units" type= "xs:token" fixed= "kV" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "c2_aperture_diameter" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_c2_aperture_diameter" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "nominal_cs" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_nominal_cs" >
<xs:attribute name= "units" type= "xs:token" fixed= "mm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "nominal_defocus_min" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_defocus_min" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "calibrated_defocus_min" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_defocus_min" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "nominal_defocus_max" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_defocus_max" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "calibrated_defocus_max" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_defocus_max" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "nominal_magnification" type= "allowed_magnification" minOccurs= "0" />
<xs:element name= "calibrated_magnification" type= "allowed_magnification" minOccurs= "0" />
<xs:element name= "specimen_holder_model" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "FISCHIONE 2550" />
<xs:enumeration value= "FEI TITAN KRIOS AUTOGRID HOLDER" />
<xs:enumeration value= "GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN 914 HIGH TILT LIQUID NITROGEN CRYO TRANSFER TOMOGRAPHY HOLDER" />
<xs:enumeration value= "GATAN 915 DOUBLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN CHDT 3504 DOUBLE TILT HIGH RESOLUTION NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN CT3500 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN CT3500TR SINGLE TILT ROTATION LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN ELSA 698 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER" />
<xs:enumeration value= "GATAN HC 3500 SINGLE TILT HEATING/NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN HCHDT 3010 DOUBLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN HCHST 3008 SINGLE TILT HIGH RESOLUTION HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN HELIUM" />
<xs:enumeration value= "GATAN LIQUID NITROGEN" />
<xs:enumeration value= "GATAN UHRST 3500 SINGLE TILT ULTRA HIGH RESOLUTION NITROGEN COOLING HOLDER" />
<xs:enumeration value= "GATAN ULTDT ULTRA LOW TEMPERATURE DOUBLE TILT HELIUM COOLING HOLDER" />
<xs:enumeration value= "GATAN ULTST ULTRA LOW TEMPERATURE SINGLE TILT HELIUM COOLING HOLDER" />
<xs:enumeration value= "HOME BUILD" />
<xs:enumeration value= "JEOL" />
<xs:enumeration value= "JEOL 3200FSC CRYOHOLDER" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PHILIPS ROTATION HOLDER" />
<xs:enumeration value= "SIDE ENTRY, EUCENTRIC" />
<xs:enumeration value= "JEOL CRYOSPECPORTER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "cooling_holder_cryogen" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "HELIUM" />
<xs:enumeration value= "NITROGEN" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "temperature" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "temperature_min" type= "temperature_type" minOccurs= "0" />
<xs:element name= "temperature_max" type= "temperature_type" minOccurs= "0" />
<xs:element name= "temperature_average" type= "temperature_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "alignment_procedure" minOccurs= "0" >
<xs:complexType >
<xs:choice >
<xs:element name= "none" >
<xs:complexType />
</xs:element>
<xs:element name= "basic" >
<xs:complexType >
<xs:sequence >
<xs:element name= "residual_tilt" type= "residual_tilt_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "zemlin_tableau" >
<xs:complexType />
</xs:element>
<xs:element name= "coma_free" >
<xs:complexType >
<xs:sequence >
<xs:element name= "residual_tilt" type= "residual_tilt_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "other" >
<xs:complexType />
</xs:element>
<xs:element name= "legacy" >
<xs:complexType >
<xs:sequence >
<xs:element name= "astigmatism" type= "xs:string" />
<xs:element name= "electron_beam_tilt_params" type= "xs:string" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:choice>
</xs:complexType>
</xs:element>
<xs:element name= "specialist_optics" type= "specialist_optics_type" minOccurs= "0" />
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "date" type= "xs:date" minOccurs= "0" />
<xs:element name= "image_recording_list" >
<xs:complexType >
<xs:sequence maxOccurs= "unbounded" >
<xs:element name= "image_recording" >
<xs:complexType >
<xs:sequence >
<xs:element name= "film_or_detector_model" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_film_or_detector_model" >
<xs:attribute name= "category" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "CCD" />
<xs:enumeration value= "CMOS" />
<xs:enumeration value= "DIRECT ELECTRON DETECTOR" />
<xs:enumeration value= "STORAGE PHOSPOR (IMAGE PLATES)" />
<xs:enumeration value= "FILM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "detector_mode" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "COUNTING" />
<xs:enumeration value= "INTEGRATING" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "SUPER-RESOLUTION" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "digitization_details" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "scanner" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EIKONIX IEEE 488" />
<xs:enumeration value= "EMIL 10" />
<xs:enumeration value= "IMACON" />
<xs:enumeration value= "NIKON COOLSCAN" />
<xs:enumeration value= "NIKON SUPER COOLSCAN 9000" />
<xs:enumeration value= "OPTRONICS" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PATCHWORK DENSITOMETER" />
<xs:enumeration value= "PERKIN ELMER" />
<xs:enumeration value= "PRIMESCAN" />
<xs:enumeration value= "TEMSCAN" />
<xs:enumeration value= "ZEISS SCAI" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "dimensions" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "width" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:positiveInteger" >
<xs:attribute name= "units" type= "xs:token" fixed= "pixel" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "height" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:positiveInteger" >
<xs:attribute name= "units" type= "xs:token" fixed= "pixel" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "sampling_interval" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_scaning_interval" >
<xs:attribute name= "units" type= "xs:token" fixed= "µm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "frames_per_image" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "number_grids_imaged" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "number_real_images" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "number_diffraction_images" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "average_exposure_time" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_average_exposure_time_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "s" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "average_electron_dose_per_image" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_electron_dose" >
<xs:attribute name= "units" type= "xs:token" fixed= "e/Å^2" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "detector_distance" type= "xs:string" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "od_range" type= "xs:float" minOccurs= "0" />
<xs:element name= "bits_per_pixel" type= "xs:float" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "image_recording_id" type= "xs:positiveInteger" />
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "specimen_holder" type= "xs:string" minOccurs= "0" />
<xs:element name= "tilt_angle_min" type= "xs:string" minOccurs= "0" />
<xs:element name= "tilt_angle_max" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "microscopy_id" type= "xs:positiveInteger" use= "required" />
</xs:complexType>
Simple Type allowed_acceleration_voltage
Simple Type allowed_c2_aperture_diameter
Simple Type allowed_nominal_cs
Simple Type allowed_defocus_min
Simple Type allowed_defocus_max
Simple Type allowed_magnification
Complex Type temperature_type
Simple Type allowed_temperature
Complex Type residual_tilt_type
Complex Type specialist_optics_type
Simple Type allowed_energy_filter_width
Simple Type allowed_energy_window
Complex Type software_list_type
Namespace No namespace
Diagram
Used by
Elements
angle_assignment_type/software_list background_masked_type/software_list base_microscopy_type/software_list classification_type/software_list crystallography_proc_add_group/lattice_distortion_correction_software_list crystallography_proc_add_group/merging_software_list crystallography_proc_add_group/symmetry_determination_software_list ctf_correction_type/software_list final_reconstruction_type/software_list layer_lines_type/helix_length/software_list layer_lines_type/indexing/software_list modelling_type/software_list molecular_replacement_type/software_list particle_selection_type/software_list reconstruction_filtering_type/b-factorSharpening/software_list reconstruction_filtering_type/other/software_list reconstruction_filtering_type/sharpening/software_list reconstruction_filtering_type/spatial_filtering/software_list refinement_type/software_list segment_selection_type/software_list single_particle_proc_add_group/final_multi_reference_alignment/software_list starting_map_type/orthogonal_tilt/software_list starting_map_type/random_conical_tilt/software_list subtomogram_averaging_proc_add_group/extraction/software_list subtomogram_averaging_proc_add_group/final_multi_reference_alignment/software_list tomography_proc_add_group/series_aligment_software_list
Model
Children software
Source
<xs:complexType name= "software_list_type" >
<xs:sequence >
<xs:element name= "software" type= "software_type" minOccurs= "0" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
Complex Type software_type
Simple Type allowed_film_or_detector_model
Namespace No namespace
Diagram
Type restriction of xs:token
Facets
enumeration
AGFA SCIENTA FILM
enumeration
DIRECT ELECTRON DE-10 (5k x 4k)
enumeration
DIRECT ELECTRON DE-12 (4k x 3k)
enumeration
DIRECT ELECTRON DE-16 (4k x 4k)
enumeration
DIRECT ELECTRON DE-20 (5k x 3k)
enumeration
DIRECT ELECTRON DE-64 (8k x 8k)
enumeration
FEI CETA (4k x 4k)
enumeration
FEI EAGLE (2k x 2k)
enumeration
FEI EAGLE (4k x 4k)
enumeration
FEI FALCON I (4k x 4k)
enumeration
FEI FALCON II (4k x 4k)
enumeration
FEI FALCON III (4k x 4k)
enumeration
FEI FALCON IV (4k x 4k)
enumeration
GATAN K2 (4k x 4k)
enumeration
GATAN K2 BASE (4k x 4k)
enumeration
GATAN K2 IS (4k x 4k)
enumeration
GATAN K2 QUANTUM (4k x 4k)
enumeration
GATAN K2 SUMMIT (4k x 4k)
enumeration
GATAN K3 (6k x 4k)
enumeration
GATAN K3 BIOQUANTUM (6k x 4k)
enumeration
GATAN MULTISCAN
enumeration
GATAN ORIUS SC1000 (4k x 2.7k)
enumeration
GATAN ORIUS SC200 (2k x 2k)
enumeration
GATAN ORIUS SC600 (2.7k x 2.7k)
enumeration
GATAN ULTRASCAN 1000 (2k x 2k)
enumeration
GATAN ULTRASCAN 10000 (10k x 10k)
enumeration
GATAN ULTRASCAN 4000 (4k x 4k)
enumeration
GENERIC CCD
enumeration
GENERIC CCD (2k x 2k)
enumeration
GENERIC CCD (4k x 4k)
enumeration
GENERIC FILM
enumeration
GENERIC GATAN
enumeration
GENERIC GATAN (2k x 2k)
enumeration
GENERIC GATAN (4k x 4k)
enumeration
GENERIC IMAGE PLATES
enumeration
GENERIC TVIPS
enumeration
GENERIC TVIPS (2k x 2k)
enumeration
GENERIC TVIPS (4k x 4k)
enumeration
KODAK 4489 FILM
enumeration
KODAK SO-163 FILM
enumeration
OTHER
enumeration
PROSCAN TEM-PIV (2k x 2k)
enumeration
SIA 15C (3k x 3k)
enumeration
TVIPS TEMCAM-F216 (2k x 2k)
enumeration
TVIPS TEMCAM-F224 (2k x 2k)
enumeration
TVIPS TEMCAM-F415 (4k x 4k)
enumeration
TVIPS TEMCAM-F416 (4k x 4k)
enumeration
TVIPS TEMCAM-F816 (8k x 8k)
Used by
Source
<xs:simpleType name= "allowed_film_or_detector_model" >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "AGFA SCIENTA FILM" />
<xs:enumeration value= "DIRECT ELECTRON DE-10 (5k x 4k)" />
<xs:enumeration value= "DIRECT ELECTRON DE-12 (4k x 3k)" />
<xs:enumeration value= "DIRECT ELECTRON DE-16 (4k x 4k)" />
<xs:enumeration value= "DIRECT ELECTRON DE-20 (5k x 3k)" />
<xs:enumeration value= "DIRECT ELECTRON DE-64 (8k x 8k)" />
<xs:enumeration value= "FEI CETA (4k x 4k)" />
<xs:enumeration value= "FEI EAGLE (2k x 2k)" />
<xs:enumeration value= "FEI EAGLE (4k x 4k)" />
<xs:enumeration value= "FEI FALCON I (4k x 4k)" />
<xs:enumeration value= "FEI FALCON II (4k x 4k)" />
<xs:enumeration value= "FEI FALCON III (4k x 4k)" />
<xs:enumeration value= "FEI FALCON IV (4k x 4k)" />
<xs:enumeration value= "GATAN K2 (4k x 4k)" />
<xs:enumeration value= "GATAN K2 BASE (4k x 4k)" />
<xs:enumeration value= "GATAN K2 IS (4k x 4k)" />
<xs:enumeration value= "GATAN K2 QUANTUM (4k x 4k)" />
<xs:enumeration value= "GATAN K2 SUMMIT (4k x 4k)" />
<xs:enumeration value= "GATAN K3 (6k x 4k)" />
<xs:enumeration value= "GATAN K3 BIOQUANTUM (6k x 4k)" />
<xs:enumeration value= "GATAN MULTISCAN" />
<xs:enumeration value= "GATAN ORIUS SC1000 (4k x 2.7k)" />
<xs:enumeration value= "GATAN ORIUS SC200 (2k x 2k)" />
<xs:enumeration value= "GATAN ORIUS SC600 (2.7k x 2.7k)" />
<xs:enumeration value= "GATAN ULTRASCAN 1000 (2k x 2k)" />
<xs:enumeration value= "GATAN ULTRASCAN 10000 (10k x 10k)" />
<xs:enumeration value= "GATAN ULTRASCAN 4000 (4k x 4k)" />
<xs:enumeration value= "GENERIC CCD" />
<xs:enumeration value= "GENERIC CCD (2k x 2k)" />
<xs:enumeration value= "GENERIC CCD (4k x 4k)" />
<xs:enumeration value= "GENERIC FILM" />
<xs:enumeration value= "GENERIC GATAN" />
<xs:enumeration value= "GENERIC GATAN (2k x 2k)" />
<xs:enumeration value= "GENERIC GATAN (4k x 4k)" />
<xs:enumeration value= "GENERIC IMAGE PLATES" />
<xs:enumeration value= "GENERIC TVIPS" />
<xs:enumeration value= "GENERIC TVIPS (2k x 2k)" />
<xs:enumeration value= "GENERIC TVIPS (4k x 4k)" />
<xs:enumeration value= "KODAK 4489 FILM" />
<xs:enumeration value= "KODAK SO-163 FILM" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "PROSCAN TEM-PIV (2k x 2k)" />
<xs:enumeration value= "SIA 15C (3k x 3k)" />
<xs:enumeration value= "TVIPS TEMCAM-F216 (2k x 2k)" />
<xs:enumeration value= "TVIPS TEMCAM-F224 (2k x 2k)" />
<xs:enumeration value= "TVIPS TEMCAM-F415 (4k x 4k)" />
<xs:enumeration value= "TVIPS TEMCAM-F416 (4k x 4k)" />
<xs:enumeration value= "TVIPS TEMCAM-F816 (8k x 8k)" />
</xs:restriction>
</xs:simpleType>
Simple Type allowed_scaning_interval
Simple Type allowed_average_exposure_time_type
Simple Type allowed_electron_dose
Complex Type base_image_processing_type
Complex Type map_type
Namespace No namespace
Diagram
Used by
Model
file ,
symmetry{0,1} ,
data_type ,
dimensions ,
origin ,
spacing ,
cell ,
axis_order ,
statistics{0,1} ,
pixel_spacing ,
contour_list{0,1} ,
label{0,1} ,
annotation_details{0,1} ,
details{0,1}
Children annotation_details axis_order cell contour_list data_type details dimensions file label origin pixel_spacing spacing statistics symmetry
Attributes
Source
<xs:complexType name= "map_type" >
<xs:sequence >
<xs:element name= "file" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "emd_\d{4,}([A-Za-z0-9_]*)\.map(\.gz|)" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "symmetry" type= "applied_symmetry_type" minOccurs= "0" />
<xs:element name= "data_type" type= "map_data_type" />
<xs:element name= "dimensions" type= "integer_vector_map_type" />
<xs:element name= "origin" >
<xs:complexType >
<xs:sequence >
<xs:element name= "col" type= "xs:integer" />
<xs:element name= "row" type= "xs:integer" />
<xs:element name= "sec" type= "xs:integer" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "spacing" >
<xs:complexType >
<xs:sequence >
<xs:element name= "x" type= "xs:positiveInteger" />
<xs:element name= "y" type= "xs:nonNegativeInteger" />
<xs:element name= "z" type= "xs:nonNegativeInteger" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "cell" >
<xs:complexType >
<xs:sequence >
<xs:element name= "a" type= "cell_type" />
<xs:element name= "b" type= "cell_type" />
<xs:element name= "c" type= "cell_type" />
<xs:element name= "alpha" type= "cell_angle_type" />
<xs:element name= "beta" type= "cell_angle_type" />
<xs:element name= "gamma" type= "cell_angle_type" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "axis_order" >
<xs:complexType >
<xs:sequence >
<xs:element name= "fast" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "X|Y|Z" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "medium" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "X|Y|Z" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "slow" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "X|Y|Z" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "statistics" type= "map_statistics_type" minOccurs= "0" />
<xs:element name= "pixel_spacing" >
<xs:complexType >
<xs:sequence >
<xs:element name= "x" type= "pixel_spacing_type" />
<xs:element name= "y" type= "pixel_spacing_type" />
<xs:element name= "z" type= "pixel_spacing_type" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "contour_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "contour" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "level" type= "xs:float" minOccurs= "0" />
<xs:element name= "source" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "EMDB" />
<xs:enumeration value= "AUTHOR" />
<xs:enumeration value= "SOFTWARE" />
<xs:enumeration value= "ANNOTATOR" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
<xs:attribute name= "primary" type= "xs:boolean" use= "required" />
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "label" type= "xs:token" minOccurs= "0" />
<xs:element name= "annotation_details" type= "xs:string" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "format" fixed= "CCP4" use= "required" />
<xs:attribute name= "size_kbytes" type= "xs:positiveInteger" use= "required" />
</xs:complexType>
Complex Type applied_symmetry_type
Simple Type point_group_symmetry_type
Complex Type helical_parameters_type
Simple Type allowed_rise_type
Simple Type allowed_twist_type
Simple Type map_data_type
Complex Type integer_vector_map_type
Complex Type cell_type
Simple Type allowed_cell_dim
Complex Type cell_angle_type
Simple Type allowed_cell_angle
Complex Type map_statistics_type
Complex Type pixel_spacing_type
Simple Type allowed_pixel_sampling
Complex Type interpretation_type
Namespace No namespace
Diagram
Used by
Model
modelling_list{0,1} ,
figure_list{0,1} ,
segmentation_list{0,1} ,
slices_list{0,1} ,
additional_map_list{0,1} ,
half_map_list{0,1}
Children additional_map_list figure_list half_map_list modelling_list segmentation_list slices_list
Source
<xs:complexType name= "interpretation_type" >
<xs:sequence >
<xs:element name= "modelling_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "modelling" type= "modelling_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "figure_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "figure" type= "figure_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "segmentation_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "segmentation" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "file" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "[emd_\d{4,}]+.*" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "mask_details" type= "map_type" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "slices_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "slice" type= "map_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "additional_map_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "additional_map" type= "map_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "half_map_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "half_map" type= "map_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<!--
<xs:element name="reference_map_for_composition" minOccurs="0">
<xs:complexType>
<xs:sequence>
<xs:element name="ref_map" minOccurs="0" type="map_type"/>
</xs:sequence>
</xs:complexType>
</xs:element>
-->
</xs:sequence>
</xs:complexType>
Complex Type modelling_type
Namespace No namespace
Diagram
Used by
Model
initial_model* ,
final_model{0,1} ,
refinement_protocol{0,1} ,
software_list{0,1} ,
details{0,1} ,
target_criteria{0,1} ,
refinement_space{0,1} ,
overall_bvalue{0,1}
Children details final_model initial_model overall_bvalue refinement_protocol refinement_space software_list target_criteria
Source
<xs:complexType name= "modelling_type" >
<xs:sequence >
<xs:element name= "initial_model" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "access_code" >
<xs:simpleType >
<xs:restriction base= "pdb_code_type" >
<xs:pattern value= "\d[\dA-Za-z]{3}" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "chain" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:complexContent >
<xs:extension base= "chain_type" >
<xs:sequence >
<xs:element name= "number_of_copies_in_final_model" type= "xs:positiveInteger" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "final_model" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "access_code" type= "pdb_code_type" />
<xs:element name= "chain" type= "chain_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "refinement_protocol" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "AB INITIO MODEL" />
<xs:enumeration value= "BACKBONE TRACE" />
<xs:enumeration value= "FLEXIBLE FIT" />
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "RIGID BODY FIT" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
<xs:element name= "target_criteria" type= "xs:token" minOccurs= "0" />
<xs:element name= "refinement_space" type= "xs:token" minOccurs= "0" />
<xs:element name= "overall_bvalue" type= "xs:float" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Complex Type chain_type
Simple Type chain_pdb_id
Complex Type figure_type
Complex Type validation_type
Simple Type author_enums
Namespace No namespace
Diagram
Type restriction of xs:string
Facets
enumeration
Accelerated Technologies Center for Gene to 3D Structure (ATCG3D)
enumeration
Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT)
enumeration
Atoms-to-Animals: The Immune Function Network (IFN)
enumeration
Bacterial targets at IGS-CNRS, France (BIGS)
enumeration
Berkeley Structural Genomics Center (BSGC)
enumeration
Center for Eukaryotic Structural Genomics (CESG)
enumeration
Center for High-Throughput Structural Biology (CHTSB)
enumeration
Center for Membrane Proteins of Infectious Diseases (MPID)
enumeration
Center for Structural Genomics of Infectious Diseases (CSGID)
enumeration
Center for Structures of Membrane Proteins (CSMP)
enumeration
Center for the X-ray Structure Determination of Human Transporters (TransportPDB)
enumeration
Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS)
enumeration
Enzyme Discovery for Natural Product Biosynthesis (NatPro)
enumeration
GPCR Network (GPCR)
enumeration
Integrated Center for Structure and Function Innovation (ISFI)
enumeration
Israel Structural Proteomics Center (ISPC)
enumeration
Joint Center for Structural Genomics (JCSG)
enumeration
Marseilles Structural Genomics Program @ AFMB (MSGP)
enumeration
Medical Structural Genomics of Pathogenic Protozoa (MSGPP)
enumeration
Membrane Protein Structural Biology Consortium (MPSBC)
enumeration
Membrane Protein Structures by Solution NMR (MPSbyNMR)
enumeration
Midwest Center for Macromolecular Research (MCMR)
enumeration
Midwest Center for Structural Genomics (MCSG)
enumeration
Mitochondrial Protein Partnership (MPP)
enumeration
Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI)
enumeration
Mycobacterium Tuberculosis Structural Proteomics Project (XMTB)
enumeration
New York Consortium on Membrane Protein Structure (NYCOMPS)
enumeration
New York SGX Research Center for Structural Genomics (NYSGXRC)
enumeration
New York Structural GenomiX Research Consortium (NYSGXRC)
enumeration
New York Structural Genomics Research Consortium (NYSGRC)
enumeration
Northeast Structural Genomics Consortium (NESG)
enumeration
Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals)
enumeration
Ontario Centre for Structural Proteomics (OCSP)
enumeration
Oxford Protein Production Facility (OPPF)
enumeration
Paris-Sud Yeast Structural Genomics (YSG)
enumeration
Partnership for Nuclear Receptor Signaling Code Biology (NHRs)
enumeration
Partnership for Stem Cell Biology (STEMCELL)
enumeration
Partnership for T-Cell Biology (TCELL)
enumeration
Program for the Characterization of Secreted Effector Proteins (PCSEP)
enumeration
Protein Structure Factory (PSF)
enumeration
QCRG Structural Biology Consortium
enumeration
RIKEN Structural Genomics/Proteomics Initiative (RSGI)
enumeration
Scottish Structural Proteomics Facility (SSPF)
enumeration
Seattle Structural Genomics Center for Infectious Disease (SSGCID)
enumeration
South Africa Structural Targets Annotation Database (SASTAD)
enumeration
Southeast Collaboratory for Structural Genomics (SECSG)
enumeration
Structural Genomics Consortium (SGC)
enumeration
Structural Genomics Consortium for Research on Gene Expression (SGCGES)
enumeration
Structural Genomics of Pathogenic Protozoa Consortium (SGPP)
enumeration
Structural Proteomics in Europe (SPINE)
enumeration
Structural Proteomics in Europe 2 (SPINE-2)
enumeration
Structure 2 Function Project (S2F)
enumeration
Structure, Dynamics and Activation Mechanisms of Chemokine Receptors (CHSAM)
enumeration
Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI)
enumeration
Structure-Function Studies of Tight Junction Membrane Proteins (TJMP)
enumeration
Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI)
enumeration
TB Structural Genomics Consortium (TBSGC)
enumeration
Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS)
enumeration
Transmembrane Protein Center (TMPC)
Source
<xs:simpleType name= "author_enums" >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "Accelerated Technologies Center for Gene to 3D Structure (ATCG3D)" />
<xs:enumeration value= "Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT)" />
<xs:enumeration value= "Atoms-to-Animals: The Immune Function Network (IFN)" />
<xs:enumeration value= "Bacterial targets at IGS-CNRS, France (BIGS)" />
<xs:enumeration value= "Berkeley Structural Genomics Center (BSGC)" />
<xs:enumeration value= "Center for Eukaryotic Structural Genomics (CESG)" />
<xs:enumeration value= "Center for High-Throughput Structural Biology (CHTSB)" />
<xs:enumeration value= "Center for Membrane Proteins of Infectious Diseases (MPID)" />
<xs:enumeration value= "Center for Structural Genomics of Infectious Diseases (CSGID)" />
<xs:enumeration value= "Center for Structures of Membrane Proteins (CSMP)" />
<xs:enumeration value= "Center for the X-ray Structure Determination of Human Transporters (TransportPDB)" />
<xs:enumeration value= "Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS)" />
<xs:enumeration value= "Enzyme Discovery for Natural Product Biosynthesis (NatPro)" />
<xs:enumeration value= "GPCR Network (GPCR)" />
<xs:enumeration value= "Integrated Center for Structure and Function Innovation (ISFI)" />
<xs:enumeration value= "Israel Structural Proteomics Center (ISPC)" />
<xs:enumeration value= "Joint Center for Structural Genomics (JCSG)" />
<xs:enumeration value= "Marseilles Structural Genomics Program @ AFMB (MSGP)" />
<xs:enumeration value= "Medical Structural Genomics of Pathogenic Protozoa (MSGPP)" />
<xs:enumeration value= "Membrane Protein Structural Biology Consortium (MPSBC)" />
<xs:enumeration value= "Membrane Protein Structures by Solution NMR (MPSbyNMR)" />
<xs:enumeration value= "Midwest Center for Macromolecular Research (MCMR)" />
<xs:enumeration value= "Midwest Center for Structural Genomics (MCSG)" />
<xs:enumeration value= "Mitochondrial Protein Partnership (MPP)" />
<xs:enumeration value= "Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI)" />
<xs:enumeration value= "Mycobacterium Tuberculosis Structural Proteomics Project (XMTB)" />
<xs:enumeration value= "New York Consortium on Membrane Protein Structure (NYCOMPS)" />
<xs:enumeration value= "New York SGX Research Center for Structural Genomics (NYSGXRC)" />
<xs:enumeration value= "New York Structural GenomiX Research Consortium (NYSGXRC)" />
<xs:enumeration value= "New York Structural Genomics Research Consortium (NYSGRC)" />
<xs:enumeration value= "Northeast Structural Genomics Consortium (NESG)" />
<xs:enumeration value= "Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals)" />
<xs:enumeration value= "Ontario Centre for Structural Proteomics (OCSP)" />
<xs:enumeration value= "Oxford Protein Production Facility (OPPF)" />
<xs:enumeration value= "Paris-Sud Yeast Structural Genomics (YSG)" />
<xs:enumeration value= "Partnership for Nuclear Receptor Signaling Code Biology (NHRs)" />
<xs:enumeration value= "Partnership for Stem Cell Biology (STEMCELL)" />
<xs:enumeration value= "Partnership for T-Cell Biology (TCELL)" />
<xs:enumeration value= "Program for the Characterization of Secreted Effector Proteins (PCSEP)" />
<xs:enumeration value= "Protein Structure Factory (PSF)" />
<xs:enumeration value= "QCRG Structural Biology Consortium" />
<xs:enumeration value= "RIKEN Structural Genomics/Proteomics Initiative (RSGI)" />
<xs:enumeration value= "Scottish Structural Proteomics Facility (SSPF)" />
<xs:enumeration value= "Seattle Structural Genomics Center for Infectious Disease (SSGCID)" />
<xs:enumeration value= "South Africa Structural Targets Annotation Database (SASTAD)" />
<xs:enumeration value= "Southeast Collaboratory for Structural Genomics (SECSG)" />
<xs:enumeration value= "Structural Genomics Consortium (SGC)" />
<xs:enumeration value= "Structural Genomics Consortium for Research on Gene Expression (SGCGES)" />
<xs:enumeration value= "Structural Genomics of Pathogenic Protozoa Consortium (SGPP)" />
<xs:enumeration value= "Structural Proteomics in Europe (SPINE)" />
<xs:enumeration value= "Structural Proteomics in Europe 2 (SPINE-2)" />
<xs:enumeration value= "Structure 2 Function Project (S2F)" />
<xs:enumeration value= "Structure, Dynamics and Activation Mechanisms of Chemokine Receptors (CHSAM)" />
<xs:enumeration value= "Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI)" />
<xs:enumeration value= "Structure-Function Studies of Tight Junction Membrane Proteins (TJMP)" />
<xs:enumeration value= "Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI)" />
<xs:enumeration value= "TB Structural Genomics Consortium (TBSGC)" />
<xs:enumeration value= "Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS)" />
<xs:enumeration value= "Transmembrane Protein Center (TMPC)" />
</xs:restriction>
</xs:simpleType>
Complex Type author_order_type
Simple Type page_type
Complex Type cell_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source*
Children category details external_references macromolecule_list name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag synthetic_source
Attributes
Source
<xs:complexType name= "cell_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "natural_source" type= "cell_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "synthetic_source" type= "cell_source_type" minOccurs= "0" maxOccurs= "unbounded" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type cell_source_type
Namespace No namespace
Diagram
Type extension of base_source_type
Type hierarchy
Used by
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:complexType name= "cell_source_type" >
<xs:complexContent >
<xs:extension base= "base_source_type" >
<xs:sequence >
<xs:element name= "organ" type= "xs:token" minOccurs= "0" />
<xs:element name= "tissue" type= "xs:token" minOccurs= "0" />
<xs:element name= "cell" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type complex_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
recombinant_expression* ,
molecular_weight{0,1} ,
ribosome-details{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag recombinant_expression ribosome-details synthetic_source
Attributes
Source
<xs:complexType name= "complex_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "natural_source" type= "complex_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "synthetic_source" type= "complex_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "recombinant_expression" type= "recombinant_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "molecular_weight" type= "molecular_weight_type" minOccurs= "0" />
<xs:element name= "ribosome-details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "chimera" type= "xs:boolean" fixed= "true" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type complex_source_type
Namespace No namespace
Diagram
Type extension of base_source_type
Type hierarchy
Used by
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:complexType name= "complex_source_type" >
<xs:complexContent >
<xs:extension base= "base_source_type" >
<xs:sequence >
<xs:element name= "organ" type= "xs:token" minOccurs= "0" />
<xs:element name= "tissue" type= "xs:token" minOccurs= "0" />
<xs:element name= "cell" type= "xs:token" minOccurs= "0" />
<xs:element name= "organelle" type= "xs:token" minOccurs= "0" />
<xs:element name= "cellular_location" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type recombinant_source_type
Complex Type organelle_or_cellular_component_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
molecular_weight{0,1} ,
recombinant_expression{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag recombinant_expression synthetic_source
Attributes
Source
<xs:complexType name= "organelle_or_cellular_component_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "natural_source" type= "organelle_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "synthetic_source" type= "organelle_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "molecular_weight" type= "molecular_weight_type" minOccurs= "0" />
<xs:element name= "recombinant_expression" type= "recombinant_source_type" minOccurs= "0" maxOccurs= "1" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type organelle_source_type
Namespace No namespace
Diagram
Type extension of base_source_type
Type hierarchy
Used by
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1} ,
organelle{0,1} ,
cellular_location{0,1}
Children cell cellular_location organ organelle organism strain synonym_organism tissue
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:complexType name= "organelle_source_type" >
<xs:complexContent >
<xs:extension base= "base_source_type" >
<xs:sequence >
<xs:element name= "organ" type= "xs:token" minOccurs= "0" />
<xs:element name= "tissue" type= "xs:token" minOccurs= "0" />
<xs:element name= "cell" type= "xs:token" minOccurs= "0" />
<xs:element name= "organelle" type= "xs:token" minOccurs= "0" />
<xs:element name= "cellular_location" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type sample_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
synthetic_source* ,
number_unique_components{0,1} ,
molecular_weight{0,1}
Children category details external_references macromolecule_list molecular_weight name natural_source number_of_copies number_unique_components oligomeric_state parent recombinant_exp_flag synthetic_source
Attributes
Source
<xs:complexType name= "sample_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "natural_source" type= "sample_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "synthetic_source" type= "sample_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "number_unique_components" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "molecular_weight" type= "molecular_weight_type" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type sample_source_type
Namespace No namespace
Diagram
Type extension of base_source_type
Type hierarchy
Used by
Model
organism ,
strain{0,1} ,
synonym_organism{0,1} ,
organ{0,1} ,
tissue{0,1} ,
cell{0,1}
Children cell organ organism strain synonym_organism tissue
Attributes
QName
Type
Use
database restriction of xs:token
optional
Source
<xs:complexType name= "sample_source_type" >
<xs:complexContent >
<xs:extension base= "base_source_type" >
<xs:sequence >
<xs:element name= "organ" type= "xs:token" minOccurs= "0" />
<xs:element name= "tissue" type= "xs:token" minOccurs= "0" />
<xs:element name= "cell" type= "xs:token" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type tissue_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
natural_source* ,
sythetic_source*
Children category details external_references macromolecule_list name natural_source number_of_copies oligomeric_state parent recombinant_exp_flag sythetic_source
Attributes
Source
<xs:complexType name= "tissue_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "natural_source" type= "tissue_source_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "sythetic_source" type= "tissue_source_type" minOccurs= "0" maxOccurs= "unbounded" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type tissue_source_type
Complex Type virus_supramolecule_type
Namespace No namespace
Diagram
Type extension of base_supramolecule_type
Type hierarchy
Used by
Model
name ,
category{0,1} ,
parent ,
macromolecule_list{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
external_references* ,
recombinant_exp_flag{0,1} ,
sci_species_name{0,1} ,
sci_species_strain{0,1} ,
natural_host* ,
synthetic_host* ,
host_system{0,1} ,
molecular_weight{0,1} ,
virus_shell* ,
virus_type ,
virus_isolate ,
virus_enveloped ,
virus_empty ,
syn_species_name{0,1} ,
sci_species_serotype{0,1} ,
sci_species_serocomplex{0,1} ,
sci_species_subspecies{0,1}
Children category details external_references host_system macromolecule_list molecular_weight name natural_host number_of_copies oligomeric_state parent recombinant_exp_flag sci_species_name sci_species_serocomplex sci_species_serotype sci_species_strain sci_species_subspecies syn_species_name synthetic_host virus_empty virus_enveloped virus_isolate virus_shell virus_type
Attributes
Source
<xs:complexType name= "virus_supramolecule_type" >
<xs:complexContent >
<xs:extension base= "base_supramolecule_type" >
<xs:sequence >
<xs:element name= "sci_species_name" type= "virus_species_name_type" minOccurs= "0" />
<xs:element name= "sci_species_strain" type= "xs:string" minOccurs= "0" maxOccurs= "1" />
<xs:element name= "natural_host" type= "virus_host_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "synthetic_host" type= "virus_host_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "host_system" type= "recombinant_source_type" minOccurs= "0" />
<xs:element name= "molecular_weight" type= "molecular_weight_type" minOccurs= "0" />
<xs:element name= "virus_shell" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "name" type= "xs:token" nillable= "false" minOccurs= "0" />
<xs:element name= "diameter" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_shell_diameter" >
<xs:attribute name= "units" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "triangulation" type= "xs:positiveInteger" minOccurs= "0" />
</xs:sequence>
<xs:attribute name= "shell_id" type= "xs:positiveInteger" />
</xs:complexType>
</xs:element>
<xs:element name= "virus_type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PRION" />
<xs:enumeration value= "SATELLITE" />
<xs:enumeration value= "VIRION" />
<xs:enumeration value= "VIROID" />
<xs:enumeration value= "VIRUS-LIKE PARTICLE" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "virus_isolate" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "OTHER" />
<xs:enumeration value= "SEROCOMPLEX" />
<xs:enumeration value= "SEROTYPE" />
<xs:enumeration value= "SPECIES" />
<xs:enumeration value= "STRAIN" />
<xs:enumeration value= "SUBSPECIES" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "virus_enveloped" type= "xs:boolean" />
<xs:element name= "virus_empty" type= "xs:boolean" />
<xs:element name= "syn_species_name" type= "xs:string" minOccurs= "0" maxOccurs= "1" />
<xs:element name= "sci_species_serotype" type= "xs:string" minOccurs= "0" maxOccurs= "1" />
<xs:element name= "sci_species_serocomplex" type= "xs:string" minOccurs= "0" maxOccurs= "1" />
<xs:element name= "sci_species_subspecies" type= "xs:string" minOccurs= "0" maxOccurs= "1" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type virus_species_name_type
Complex Type virus_host_type
Simple Type allowed_shell_diameter
Complex Type dna_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence ,
classification{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1}
Children classification details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag sequence structure synthetic_flag synthetic_source
Attributes
Source
<xs:complexType name= "dna_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "sequence" >
<xs:complexType >
<xs:sequence >
<xs:element name= "string" type= "xs:token" />
<xs:element name= "discrepancy_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "discrepancy" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "[ ARNDCEQGHILKMFPSTWYVUOBZJX\(\)]\d+[ ARNDCEQGHILKMFPSTWYVUOBZJX\(\)]" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "REFSEQ" />
<xs:enumeration value= "GENBANK" />
<xs:whiteSpace value= "collapse" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "classification" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "DNA" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "structure" type= "xs:token" minOccurs= "0" />
<xs:element name= "synthetic_flag" type= "xs:boolean" minOccurs= "0" />
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type em_label_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
formula{0,1} ,
synthetic_source{0,1}
Children details formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag synthetic_source
Attributes
Source
<xs:complexType name= "em_label_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "formula" type= "formula_type" minOccurs= "0" />
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type ligand_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
formula{0,1} ,
external_references* ,
recombinant_expression{0,1}
Children details external_references formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression
Attributes
Source
<xs:complexType name= "ligand_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "formula" type= "formula_type" minOccurs= "0" />
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CAS" />
<xs:enumeration value= "PUBCHEM" />
<xs:enumeration value= "DRUGBANK" />
<xs:enumeration value= "CHEBI" />
<xs:enumeration value= "CHEMBL" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "recombinant_expression" type= "recombinant_source_type" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type other_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence{0,1} ,
classification ,
recombinant_expression{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1}
Children classification details molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression sequence structure synthetic_flag synthetic_source
Attributes
Source
<xs:complexType name= "other_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "sequence" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "string" >
<xs:simpleType >
<xs:restriction base= "xs:token" />
</xs:simpleType>
</xs:element>
<xs:element name= "discrepancy_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "discrepancy" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "[AGCTRYSWKMBDHVN\.-]\d+[AGCTRYSWKMBDHVN\.-]" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" type= "xs:token" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "classification" type= "xs:token" />
<xs:element name= "recombinant_expression" type= "recombinant_source_type" minOccurs= "0" />
<xs:element name= "structure" type= "xs:token" minOccurs= "0" />
<xs:element name= "synthetic_flag" type= "xs:boolean" minOccurs= "0" />
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type protein_or_peptide_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
recombinant_expression{0,1} ,
synthetic_source{0,1} ,
enantiomer ,
sequence ,
ec_number*
Children details ec_number enantiomer molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag recombinant_expression sequence synthetic_source
Attributes
Source
<xs:complexType name= "protein_or_peptide_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "recombinant_expression" type= "recombinant_source_type" minOccurs= "0" />
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
<xs:element name= "enantiomer" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "LEVO" />
<xs:enumeration value= "DEXTRO" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "sequence" >
<xs:complexType >
<xs:sequence >
<xs:element name= "string" type= "xs:token" minOccurs= "0" />
<xs:element name= "discrepancy_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "discrepancy" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "[ARNDCEQGHILKMFPSTWYVUOBZJX]\d+[ARNDCEQGHILKMFPSTWYVUOBZJX]" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "connectivity" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element minOccurs= "0" name= "_n-link" >
<xs:complexType >
<xs:sequence >
<xs:element name= "molecule_id" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element minOccurs= "0" name= "_c-link" >
<xs:complexType >
<xs:sequence >
<xs:element name= "molecule_id" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "UNIPROTKB" />
<xs:enumeration value= "UNIPARC" />
<xs:enumeration value= "INTERPRO" />
<xs:enumeration value= "GO" />
<xs:enumeration value= "GENBANK" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "ec_number" minOccurs= "0" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "([1-7]((.[1-9][0-9]?)|(.-))((.[1-9][0-9]?)|(.-))((.[1-9][0-9]?[0-9]?)|(.-)))(([ ]*,[ ]*)([1-6]((.[1-9][0-9]?)|(.-))((.[1-9][0-9]?)|(.-))((.[1-9][0-9]?[0-9]?)|(.-))))*" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type rna_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
sequence ,
classification{0,1} ,
structure{0,1} ,
synthetic_flag{0,1} ,
synthetic_source{0,1} ,
ec_number*
Children classification details ec_number molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag sequence structure synthetic_flag synthetic_source
Attributes
Source
<xs:complexType name= "rna_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "sequence" >
<xs:complexType >
<xs:sequence >
<xs:element name= "string" type= "xs:token" />
<xs:element name= "discrepancy_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "discrepancy" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "[ ARNDCEQGHILKMFPSTWYVUOBZJX\(\)]\d+[ ARNDCEQGHILKMFPSTWYVUOBZJX\(\)]" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "REFSEQ" />
<xs:enumeration value= "GENBANK" />
<xs:enumeration value= "UNIPROTKB" />
<xs:whiteSpace value= "collapse" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "classification" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "MESSENGER" />
<xs:enumeration value= "TRANSFER" />
<xs:enumeration value= "RIBOSOMAL" />
<xs:enumeration value= "NON-CODING" />
<xs:enumeration value= "INTERFERENCE" />
<xs:enumeration value= "SMALL INTERFERENCE" />
<xs:enumeration value= "GENOMIC" />
<xs:enumeration value= "PRE-MESSENGER" />
<xs:enumeration value= "SMALL NUCLEOLAR" />
<xs:enumeration value= "TRANSFER-MESSENGER" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "structure" type= "xs:token" minOccurs= "0" />
<xs:element name= "synthetic_flag" type= "xs:boolean" minOccurs= "0" />
<xs:element name= "synthetic_source" type= "macromolecule_source_type" minOccurs= "0" />
<xs:element name= "ec_number" minOccurs= "0" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:pattern value= "\d+(\.(\d+|\-)){3}" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type saccharide_macromolecule_type
Namespace No namespace
Diagram
Type extension of base_macromolecule_type
Type hierarchy
Used by
Model
name ,
natural_source{0,1} ,
molecular_weight{0,1} ,
details{0,1} ,
number_of_copies{0,1} ,
oligomeric_state{0,1} ,
recombinant_exp_flag{0,1} ,
enantiomer ,
formula{0,1} ,
external_references*
Children details enantiomer external_references formula molecular_weight name natural_source number_of_copies oligomeric_state recombinant_exp_flag
Attributes
Source
<xs:complexType name= "saccharide_macromolecule_type" >
<xs:complexContent >
<xs:extension base= "base_macromolecule_type" >
<xs:sequence >
<xs:element name= "enantiomer" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "LEVO" />
<xs:enumeration value= "DEXTRO" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "formula" type= "formula_type" minOccurs= "0" />
<xs:element name= "external_references" minOccurs= "0" maxOccurs= "unbounded" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "xs:token" >
<xs:attribute name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "CARDBANK" />
<xs:whiteSpace value= "collapse" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type crystallography_preparation_type
Namespace No namespace
Diagram
Type extension of base_preparation_type
Type hierarchy
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1} ,
crystal_formation{0,1}
Children buffer concentration crystal_formation details grid shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "crystallography_preparation_type" >
<xs:complexContent >
<xs:extension base= "base_preparation_type" >
<xs:sequence >
<xs:element name= "crystal_formation" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "lipid_protein_ratio" type= "xs:float" minOccurs= "0" />
<xs:element name= "lipid_mixture" type= "xs:token" minOccurs= "0" />
<xs:element name= "instrument" type= "xs:token" minOccurs= "0" />
<xs:element name= "atmosphere" type= "xs:token" minOccurs= "0" />
<xs:element name= "temperature" type= "crystal_formation_temperature_type" minOccurs= "0" />
<xs:element name= "time" type= "crystal_formation_time_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type crystal_formation_temperature_type
Simple Type allowed_crystal_formation_temperature_type
Complex Type crystal_formation_time_type
Simple Type non_zero_float
Complex Type helical_preparation_type
Namespace No namespace
Diagram
Type extension of base_preparation_type
Type hierarchy
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "helical_preparation_type" >
<xs:complexContent >
<xs:extension base= "base_preparation_type" />
</xs:complexContent>
</xs:complexType>
Complex Type single_particle_preparation_type
Namespace No namespace
Diagram
Type extension of base_preparation_type
Type hierarchy
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "single_particle_preparation_type" >
<xs:complexContent >
<xs:extension base= "base_preparation_type" />
</xs:complexContent>
</xs:complexType>
Complex Type subtomogram_averaging_preparation_type
Namespace No namespace
Diagram
Type extension of base_preparation_type
Type hierarchy
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1}
Children buffer concentration details grid shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "subtomogram_averaging_preparation_type" >
<xs:complexContent >
<xs:extension base= "base_preparation_type" />
</xs:complexContent>
</xs:complexType>
Complex Type tomography_preparation_type
Namespace No namespace
Diagram
Type extension of base_preparation_type
Type hierarchy
Used by
Model
concentration{0,1} ,
buffer{0,1} ,
staining{0,1} ,
sugar_embedding{0,1} ,
shadowing{0,1} ,
grid{0,1} ,
vitrification{0,1} ,
details{0,1} ,
fiducial_markers_list{0,1} ,
high_pressure_freezing{0,1} ,
embedding_material{0,1} ,
cryo_protectant{0,1} ,
sectioning{0,1}
Children buffer concentration cryo_protectant details embedding_material fiducial_markers_list grid high_pressure_freezing sectioning shadowing staining sugar_embedding vitrification
Attributes
Source
<xs:complexType name= "tomography_preparation_type" >
<xs:complexContent >
<xs:extension base= "base_preparation_type" >
<xs:sequence >
<xs:element name= "fiducial_markers_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "fiducial_marker" type= "fiducial_marker_type" maxOccurs= "unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "high_pressure_freezing" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "instrument" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "BAL-TEC HPM 010" />
<xs:enumeration value= "EMS-002 RAPID IMMERSION FREEZER" />
<xs:enumeration value= "LEICA EM HPM100" />
<xs:enumeration value= "LEICA EM PACT" />
<xs:enumeration value= "LEICA EM PACT2" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "embedding_material" type= "xs:token" minOccurs= "0" />
<xs:element name= "cryo_protectant" type= "xs:token" minOccurs= "0" />
<xs:element name= "sectioning" minOccurs= "0" >
<xs:complexType >
<xs:choice >
<xs:element name= "ultramicrotomy" >
<xs:complexType >
<xs:sequence >
<xs:element name= "instrument" type= "xs:token" />
<xs:element name= "temperature" type= "temperature_type" />
<xs:element name= "final_thickness" type= "ultramicrotomy_final_thickness_type" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "focused_ion_beam" >
<xs:complexType >
<xs:sequence >
<xs:element name= "instrument" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "DB235" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "ion" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "GALLIUM+" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "voltage" type= "fib_voltage_type" />
<xs:element name= "current" type= "fib_current_type" />
<xs:element name= "dose_rate" type= "fib_dose_rate_type" minOccurs= "0" />
<xs:element name= "duration" type= "fib_duration_type" />
<xs:element name= "temperature" type= "temperature_type" />
<xs:element name= "initial_thickness" type= "fib_initial_thickness_type" />
<xs:element name= "final_thickness" type= "fib_final_thickness_type" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "other_sectioning" type= "xs:string" />
</xs:choice>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type fiducial_marker_type
Complex Type fiducial_marker_diameter_type
Simple Type allowed_diameter_colloidal_gold
Complex Type ultramicrotomy_final_thickness_type
Simple Type allowed_microtome_thickness
Complex Type fib_voltage_type
Simple Type allowed_focus_ion_voltage
Complex Type fib_current_type
Simple Type allowed_focus_ion_current
Complex Type fib_dose_rate_type
Simple Type allowed_focus_ion_dose_rate
Complex Type fib_duration_type
Complex Type fib_initial_thickness_type
Simple Type allowed_focus_ion_initial_thickness
Complex Type fib_final_thickness_type
Simple Type allowed_focus_ion_final_thickness
Complex Type crystallography_microscopy_type
Namespace No namespace
Diagram
Type extension of base_microscopy_type
Type hierarchy
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1} ,
camera_length , (
tilt_list{0,1} |
tilt_series* )
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification camera_length cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min tilt_list tilt_series
Attributes
Source
<xs:complexType name= "crystallography_microscopy_type" >
<xs:complexContent >
<xs:extension base= "base_microscopy_type" >
<xs:sequence >
<xs:element name= "camera_length" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_camera_length" >
<xs:attribute name= "units" type= "xs:token" fixed= "mm" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:choice >
<xs:element name= "tilt_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "angle" maxOccurs= "unbounded" >
<xs:simpleType >
<xs:restriction base= "xs:float" >
<xs:minInclusive value= "-70" />
<xs:maxInclusive value= "70" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "tilt_series" type= "tilt_series_type" minOccurs= "0" maxOccurs= "unbounded" />
</xs:choice>
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Simple Type allowed_camera_length
Complex Type tilt_series_type
Complex Type axis_type
Simple Type allowed_angle_tomography
Simple Type allowed_angle_increment
Complex Type helical_microscopy_type
Namespace No namespace
Diagram
Type extension of base_microscopy_type
Type hierarchy
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Attributes
Source
<xs:complexType name= "helical_microscopy_type" >
<xs:complexContent >
<xs:extension base= "base_microscopy_type" />
</xs:complexContent>
</xs:complexType>
Complex Type single_particle_microscopy_type
Namespace No namespace
Diagram
Type extension of base_microscopy_type
Type hierarchy
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1}
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min
Attributes
Source
<xs:complexType name= "single_particle_microscopy_type" >
<xs:complexContent >
<xs:extension base= "base_microscopy_type" />
</xs:complexContent>
</xs:complexType>
Complex Type tomography_microscopy_type
Namespace No namespace
Diagram
Type extension of base_microscopy_type
Type hierarchy
Used by
Model
specimen_preparations{0,1} ,
microscope ,
illumination_mode ,
imaging_mode ,
electron_source ,
acceleration_voltage ,
c2_aperture_diameter{0,1} ,
nominal_cs{0,1} ,
nominal_defocus_min{0,1} ,
calibrated_defocus_min{0,1} ,
nominal_defocus_max{0,1} ,
calibrated_defocus_max{0,1} ,
nominal_magnification{0,1} ,
calibrated_magnification{0,1} ,
specimen_holder_model{0,1} ,
cooling_holder_cryogen{0,1} ,
temperature{0,1} ,
alignment_procedure{0,1} ,
specialist_optics{0,1} ,
software_list{0,1} ,
details{0,1} ,
date{0,1} ,
image_recording_list ,
specimen_holder{0,1} ,
tilt_angle_min{0,1} ,
tilt_angle_max{0,1} ,
tilt_series*
Children acceleration_voltage alignment_procedure c2_aperture_diameter calibrated_defocus_max calibrated_defocus_min calibrated_magnification cooling_holder_cryogen date details electron_source illumination_mode image_recording_list imaging_mode microscope nominal_cs nominal_defocus_max nominal_defocus_min nominal_magnification software_list specialist_optics specimen_holder specimen_holder_model specimen_preparations temperature tilt_angle_max tilt_angle_min tilt_series
Attributes
Source
<xs:complexType name= "tomography_microscopy_type" >
<xs:complexContent >
<xs:extension base= "base_microscopy_type" >
<xs:sequence >
<xs:element name= "tilt_series" type= "tilt_series_type" minOccurs= "0" maxOccurs= "unbounded" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type crystallography_processing_type
Namespace No namespace
Diagram
Type extension of base_image_processing_type
Type hierarchy
Used by
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
crystal_parameters{0,1} ,
startup_model* ,
ctf_correction{0,1} ,
molecular_replacement{0,1} ,
lattice_distortion_correction_software_list{0,1} ,
symmetry_determination_software_list{0,1} ,
merging_software_list{0,1} ,
crystallography_statistics{0,1}
Children crystal_parameters crystallography_statistics ctf_correction details final_reconstruction image_recording_id lattice_distortion_correction_software_list merging_software_list molecular_replacement startup_model symmetry_determination_software_list
Attributes
Source
<xs:complexType name= "crystallography_processing_type" >
<xs:complexContent >
<xs:extension base= "base_image_processing_type" >
<xs:group ref= "crystallography_proc_add_group" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type non_subtom_final_reconstruction_type
Namespace No namespace
Diagram
Type extension of final_reconstruction_type
Type hierarchy
Used by
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_images_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_images_used reconstruction_filtering resolution resolution_method software_list
Source
<xs:complexType name= "non_subtom_final_reconstruction_type" >
<xs:complexContent >
<xs:extension base= "final_reconstruction_type" >
<xs:sequence >
<xs:element name= "number_images_used" type= "xs:positiveInteger" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type final_reconstruction_type
Namespace No namespace
Diagram
Used by
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1}
Children algorithm applied_symmetry details number_classes_used reconstruction_filtering resolution resolution_method software_list
Source
<xs:complexType name= "final_reconstruction_type" >
<xs:sequence >
<xs:element name= "number_classes_used" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "applied_symmetry" type= "applied_symmetry_type" minOccurs= "0" />
<xs:element name= "algorithm" type= "reconstruction_algorithm_type" minOccurs= "0" />
<xs:element name= "resolution" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
<xs:attribute name= "res_type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "BY AUTHOR" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "resolution_method" minOccurs= "0" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "DIFFRACTION PATTERN/LAYERLINES" />
<xs:enumeration value= "FSC 0.143 CUT-OFF" />
<xs:enumeration value= "FSC 0.33 CUT-OFF" />
<xs:enumeration value= "FSC 0.5 CUT-OFF" />
<xs:enumeration value= "FSC 1/2 BIT CUT-OFF" />
<xs:enumeration value= "FSC 3 SIGMA CUT-OFF" />
<xs:enumeration value= "OTHER" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name= "reconstruction_filtering" type= "reconstruction_filtering_type" minOccurs= "0" />
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Simple Type reconstruction_algorithm_type
Namespace No namespace
Diagram
Type restriction of xs:token
Facets
enumeration
ALGEBRAIC (ARTS)
enumeration
BACK PROJECTION
enumeration
EXACT BACK PROJECTION
enumeration
FOURIER SPACE
enumeration
SIMULTANEOUS ITERATIVE (SIRT)
Used by
Source
<xs:simpleType name= "reconstruction_algorithm_type" >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "ALGEBRAIC (ARTS)" />
<xs:enumeration value= "BACK PROJECTION" />
<xs:enumeration value= "EXACT BACK PROJECTION" />
<xs:enumeration value= "FOURIER SPACE" />
<xs:enumeration value= "SIMULTANEOUS ITERATIVE (SIRT)" />
</xs:restriction>
</xs:simpleType>
Simple Type resolution_type
Complex Type reconstruction_filtering_type
Complex Type background_masked_type
Simple Type allowed_brestore_type
Complex Type crystal_parameters_type
Complex Type unit_cell_type
Complex Type starting_map_type
Simple Type allowed_tilt_angle_random_conical
Simple Type allowed_tilt_angle1Orthogonal
Simple Type allowed_tilt_angle2Orthogonal
Complex Type pdb_model_type
Complex Type ctf_correction_type
Simple Type correction_space_type
Complex Type molecular_replacement_type
Complex Type crystallography_statistics_type
Namespace No namespace
Diagram
Used by
Model
number_intensities_measured ,
number_structure_factors ,
fourier_space_coverage ,
r_sym{0,1} ,
r_merge ,
overall_phase_error{0,1} ,
overall_phase_residual{0,1} ,
phase_error_rejection_criteria ,
high_resolution ,
shell_list{0,1} ,
details{0,1}
Children details fourier_space_coverage high_resolution number_intensities_measured number_structure_factors overall_phase_error overall_phase_residual phase_error_rejection_criteria r_merge r_sym shell_list
Source
<xs:complexType name= "crystallography_statistics_type" >
<xs:sequence >
<xs:element name= "number_intensities_measured" type= "xs:positiveInteger" />
<xs:element name= "number_structure_factors" type= "xs:positiveInteger" />
<xs:element name= "fourier_space_coverage" type= "xs:float" />
<xs:element name= "r_sym" type= "xs:float" minOccurs= "0" />
<xs:element name= "r_merge" type= "xs:float" />
<xs:element name= "overall_phase_error" type= "xs:token" minOccurs= "0" />
<xs:element name= "overall_phase_residual" type= "xs:float" minOccurs= "0" />
<xs:element name= "phase_error_rejection_criteria" type= "xs:token" />
<xs:element name= "high_resolution" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "shell_list" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "shell" maxOccurs= "unbounded" >
<xs:complexType >
<xs:sequence >
<xs:element name= "high_resolution" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "low_resolution" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "resolution_type" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "number_structure_factors" type= "xs:positiveInteger" />
<xs:element name= "phase_residual" type= "xs:float" />
<xs:element name= "fourier_space_coverage" type= "xs:float" />
<xs:element name= "multiplicity" type= "xs:float" />
</xs:sequence>
<xs:attribute name= "shell_id" type= "xs:positiveInteger" />
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Complex Type helical_processing_type
Namespace No namespace
Diagram
Type extension of base_image_processing_type
Type hierarchy
Used by
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
ctf_correction{0,1} ,
segment_selection* ,
refinement{0,1} ,
startup_model* ,
helical_layer_lines{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details final_angle_assignment final_reconstruction helical_layer_lines image_recording_id initial_angle_assignment refinement segment_selection startup_model
Attributes
Source
<xs:complexType name= "helical_processing_type" >
<xs:complexContent >
<xs:extension base= "base_image_processing_type" >
<xs:group ref= "helical_processing_add_group" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type segment_selection_type
Namespace No namespace
Diagram
Used by
Model
number_selected{0,1} ,
segment_length{0,1} ,
segment_overlap{0,1} ,
total_filament_length{0,1} ,
software_list{0,1} ,
details{0,1}
Children details number_selected segment_length segment_overlap software_list total_filament_length
Source
<xs:complexType name= "segment_selection_type" >
<xs:sequence >
<xs:element name= "number_selected" type= "xs:positiveInteger" minOccurs= "0" />
<xs:element name= "segment_length" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "non_zero_float" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "segment_overlap" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "non_zero_float" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "total_filament_length" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "non_zero_float" >
<xs:attribute name= "units" type= "xs:token" fixed= "Å" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
Complex Type refinement_type
Complex Type layer_lines_type
Complex Type angle_assignment_type
Simple Type allowed_angular_sampling
Complex Type singleparticle_processing_type
Namespace No namespace
Diagram
Type extension of base_image_processing_type
Type hierarchy
Used by
Model
image_recording_id ,
details{0,1} ,
particle_selection* ,
ctf_correction{0,1} ,
startup_model* ,
final_reconstruction{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
final_multi_reference_alignment{0,1} ,
final_two_d_classification{0,1} ,
final_three_d_classification{0,1}
Children ctf_correction details final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification final_two_d_classification image_recording_id initial_angle_assignment particle_selection startup_model
Attributes
Source
<xs:complexType name= "singleparticle_processing_type" >
<xs:complexContent >
<xs:extension base= "base_image_processing_type" >
<xs:group ref= "single_particle_proc_add_group" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type particle_selection_type
Complex Type classification_type
Complex Type subtomogram_averaging_processing_type
Namespace No namespace
Diagram
Type extension of base_image_processing_type
Type hierarchy
Used by
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
extraction ,
ctf_correction{0,1} ,
final_multi_reference_alignment{0,1} ,
final_three_d_classification{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details extraction final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification image_recording_id
Attributes
Source
<xs:complexType name= "subtomogram_averaging_processing_type" >
<xs:complexContent >
<xs:extension base= "base_image_processing_type" >
<xs:group ref= "subtomogram_averaging_proc_add_group" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type subtomogram_final_reconstruction_type
Namespace No namespace
Diagram
Type extension of final_reconstruction_type
Type hierarchy
Used by
Model
number_classes_used{0,1} ,
applied_symmetry{0,1} ,
algorithm{0,1} ,
resolution{0,1} ,
resolution_method{0,1} ,
reconstruction_filtering{0,1} ,
software_list{0,1} ,
details{0,1} ,
number_subtomograms_used{0,1}
Children algorithm applied_symmetry details number_classes_used number_subtomograms_used reconstruction_filtering resolution resolution_method software_list
Source
<xs:complexType name= "subtomogram_final_reconstruction_type" >
<xs:complexContent >
<xs:extension base= "final_reconstruction_type" >
<xs:sequence >
<xs:element name= "number_subtomograms_used" type= "xs:positiveInteger" minOccurs= "0" />
</xs:sequence>
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type tomography_processing_type
Namespace No namespace
Diagram
Type extension of base_image_processing_type
Type hierarchy
Used by
Model
image_recording_id ,
details{0,1} ,
final_reconstruction{0,1} ,
series_aligment_software_list{0,1} ,
ctf_correction{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction details final_reconstruction image_recording_id series_aligment_software_list
Attributes
Source
<xs:complexType name= "tomography_processing_type" >
<xs:complexContent >
<xs:extension base= "base_image_processing_type" >
<xs:group ref= "tomography_proc_add_group" />
</xs:extension>
</xs:complexContent>
</xs:complexType>
Complex Type crystallography_validation_type
Complex Type fsc_curve_validation_type
Complex Type layer_lines_validation_type
Complex Type structure_factors_validation_type
Namespace No namespace
Type restriction of xs:token
Properties
Facets
enumeration
UNIPROTKB
enumeration
UNIPARC
enumeration
INTERPRO
enumeration
GO
Used by
Source
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "UNIPROTKB" />
<xs:enumeration value= "UNIPARC" />
<xs:enumeration value= "INTERPRO" />
<xs:enumeration value= "GO" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:string
Properties
Facets
enumeration
CCD
enumeration
CMOS
enumeration
DIRECT ELECTRON DETECTOR
enumeration
STORAGE PHOSPOR (IMAGE PLATES)
enumeration
FILM
Used by
Source
<xs:attribute name= "category" >
<xs:simpleType >
<xs:restriction base= "xs:string" >
<xs:enumeration value= "CCD" />
<xs:enumeration value= "CMOS" />
<xs:enumeration value= "DIRECT ELECTRON DETECTOR" />
<xs:enumeration value= "STORAGE PHOSPOR (IMAGE PLATES)" />
<xs:enumeration value= "FILM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:token
Properties
Facets
whiteSpace
collapse
enumeration
PUBMED
enumeration
DOI
enumeration
ISBN
enumeration
ISSN
enumeration
CAS
enumeration
CSD
enumeration
MEDLINE
enumeration
ASTM
Used by
Source
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PUBMED" />
<xs:enumeration value= "DOI" />
<xs:enumeration value= "ISBN" />
<xs:enumeration value= "ISSN" />
<xs:enumeration value= "CAS" />
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CSD" />
<xs:enumeration value= "MEDLINE" />
<xs:enumeration value= "ASTM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:token
Properties
Facets
whiteSpace
collapse
enumeration
PUBMED
enumeration
DOI
enumeration
ISBN
enumeration
ISSN
enumeration
CAS
enumeration
CSD
enumeration
MEDLINE
enumeration
ASTM
Used by
Source
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "PUBMED" />
<xs:enumeration value= "DOI" />
<xs:enumeration value= "ISBN" />
<xs:enumeration value= "ISSN" />
<xs:enumeration value= "CAS" />
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CSD" />
<xs:enumeration value= "MEDLINE" />
<xs:enumeration value= "ASTM" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:token
Properties
Facets
whiteSpace
collapse
enumeration
CAS
enumeration
PUBCHEM
enumeration
DRUGBANK
enumeration
CHEBI
enumeration
CHEMBL
Used by
Source
<xs:attribute name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:whiteSpace value= "collapse" />
<xs:enumeration value= "CAS" />
<xs:enumeration value= "PUBCHEM" />
<xs:enumeration value= "DRUGBANK" />
<xs:enumeration value= "CHEBI" />
<xs:enumeration value= "CHEMBL" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:token
Properties
Facets
enumeration
UNIPROTKB
enumeration
UNIPARC
enumeration
INTERPRO
enumeration
GO
enumeration
GENBANK
Used by
Source
<xs:attribute name= "type" use= "required" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "UNIPROTKB" />
<xs:enumeration value= "UNIPARC" />
<xs:enumeration value= "INTERPRO" />
<xs:enumeration value= "GO" />
<xs:enumeration value= "GENBANK" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Namespace No namespace
Type restriction of xs:token
Properties
Facets
whiteSpace
collapse
enumeration
REFSEQ
enumeration
GENBANK
enumeration
UNIPROTKB
Used by
Source
<xs:attribute name= "type" >
<xs:simpleType >
<xs:restriction base= "xs:token" >
<xs:enumeration value= "REFSEQ" />
<xs:enumeration value= "GENBANK" />
<xs:enumeration value= "UNIPROTKB" />
<xs:whiteSpace value= "collapse" />
</xs:restriction>
</xs:simpleType>
</xs:attribute>
Element Group crystallography_proc_add_group
Namespace No namespace
Diagram
Used by
Model
final_reconstruction{0,1} ,
crystal_parameters{0,1} ,
startup_model* ,
ctf_correction{0,1} ,
molecular_replacement{0,1} ,
lattice_distortion_correction_software_list{0,1} ,
symmetry_determination_software_list{0,1} ,
merging_software_list{0,1} ,
crystallography_statistics{0,1}
Children crystal_parameters crystallography_statistics ctf_correction final_reconstruction lattice_distortion_correction_software_list merging_software_list molecular_replacement startup_model symmetry_determination_software_list
Source
<xs:group name= "crystallography_proc_add_group" >
<xs:sequence >
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
<xs:element name= "crystal_parameters" type= "crystal_parameters_type" minOccurs= "0" />
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "ctf_correction" type= "ctf_correction_type" minOccurs= "0" />
<xs:element name= "molecular_replacement" type= "molecular_replacement_type" minOccurs= "0" />
<xs:element name= "lattice_distortion_correction_software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "symmetry_determination_software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "merging_software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "crystallography_statistics" type= "crystallography_statistics_type" minOccurs= "0" />
</xs:sequence>
</xs:group>
Element Group helical_processing_add_group
Namespace No namespace
Diagram
Used by
Model
final_reconstruction{0,1} ,
ctf_correction{0,1} ,
segment_selection* ,
refinement{0,1} ,
startup_model* ,
helical_layer_lines{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction final_angle_assignment final_reconstruction helical_layer_lines initial_angle_assignment refinement segment_selection startup_model
Source
<xs:group name= "helical_processing_add_group" >
<xs:sequence >
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
<xs:element name= "ctf_correction" type= "ctf_correction_type" minOccurs= "0" />
<xs:element name= "segment_selection" type= "segment_selection_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "refinement" type= "refinement_type" minOccurs= "0" />
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "helical_layer_lines" type= "layer_lines_type" minOccurs= "0" />
<xs:element name= "initial_angle_assignment" type= "angle_assignment_type" minOccurs= "0" />
<xs:element name= "final_angle_assignment" type= "angle_assignment_type" minOccurs= "0" />
<xs:element name= "crystal_parameters" type= "crystal_parameters_type" minOccurs= "0" />
</xs:sequence>
</xs:group>
Element Group single_particle_proc_add_group
Namespace No namespace
Diagram
Used by
Model
particle_selection* ,
ctf_correction{0,1} ,
startup_model* ,
final_reconstruction{0,1} ,
initial_angle_assignment{0,1} ,
final_angle_assignment{0,1} ,
final_multi_reference_alignment{0,1} ,
final_two_d_classification{0,1} ,
final_three_d_classification{0,1}
Children ctf_correction final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification final_two_d_classification initial_angle_assignment particle_selection startup_model
Source
<xs:group name= "single_particle_proc_add_group" >
<xs:sequence >
<xs:element name= "particle_selection" type= "particle_selection_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "ctf_correction" type= "ctf_correction_type" minOccurs= "0" />
<xs:element name= "startup_model" type= "starting_map_type" minOccurs= "0" maxOccurs= "unbounded" />
<xs:element name= "final_reconstruction" type= "non_subtom_final_reconstruction_type" minOccurs= "0" />
<xs:element name= "initial_angle_assignment" type= "angle_assignment_type" minOccurs= "0" />
<xs:element name= "final_angle_assignment" type= "angle_assignment_type" minOccurs= "0" />
<xs:element name= "final_multi_reference_alignment" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "number_reference_projections" type= "xs:positiveInteger" />
<xs:element name= "merit_function" minOccurs= "0" type= "xs:string" />
<xs:element name= "angular_sampling" minOccurs= "0" >
<xs:complexType >
<xs:simpleContent >
<xs:extension base= "allowed_angular_sampling" >
<xs:attribute name= "units" type= "xs:token" fixed= "degrees" use= "required" />
</xs:extension>
</xs:simpleContent>
</xs:complexType>
</xs:element>
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "final_two_d_classification" type= "classification_type" minOccurs= "0" />
<xs:element name= "final_three_d_classification" type= "classification_type" minOccurs= "0" />
</xs:sequence>
</xs:group>
Element Group subtomogram_averaging_proc_add_group
Namespace No namespace
Diagram
Used by
Model
final_reconstruction{0,1} ,
extraction ,
ctf_correction{0,1} ,
final_multi_reference_alignment{0,1} ,
final_three_d_classification{0,1} ,
final_angle_assignment{0,1} ,
crystal_parameters{0,1}
Children crystal_parameters ctf_correction extraction final_angle_assignment final_multi_reference_alignment final_reconstruction final_three_d_classification
Source
<xs:group name= "subtomogram_averaging_proc_add_group" >
<xs:sequence >
<xs:element name= "final_reconstruction" type= "subtomogram_final_reconstruction_type" minOccurs= "0" />
<xs:element name= "extraction" >
<xs:complexType >
<xs:sequence >
<xs:element name= "number_tomograms" type= "xs:positiveInteger" />
<xs:element name= "number_images_used" type= "xs:positiveInteger" />
<xs:element name= "reference_model" type= "xs:token" minOccurs= "0" />
<xs:element name= "method" type= "xs:string" minOccurs= "0" />
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "ctf_correction" type= "ctf_correction_type" minOccurs= "0" />
<xs:element name= "final_multi_reference_alignment" minOccurs= "0" >
<xs:complexType >
<xs:sequence >
<xs:element name= "number_reference_projections" type= "xs:positiveInteger" />
<xs:element name= "merit_function" minOccurs= "0" type= "xs:string" />
<xs:element name= "software_list" type= "software_list_type" minOccurs= "0" />
<xs:element name= "details" type= "xs:string" minOccurs= "0" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name= "final_three_d_classification" type= "classification_type" minOccurs= "0" />
<xs:element name= "final_angle_assignment" type= "angle_assignment_type" minOccurs= "0" />
<xs:element name= "crystal_parameters" type= "crystal_parameters_type" minOccurs= "0" />
</xs:sequence>
</xs:group>
Element Group tomography_proc_add_group